3PKO
 
 | | Crystal structure of geranylgeranyl pyrophosphate synthase from lactobacillus brevis atcc 367 complexed with citrate | | Descriptor: | CITRIC ACID, GLYCEROL, Geranylgeranyl pyrophosphate synthase | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Chang, S, Sauder, J.M, Poulter, C.D, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-11-11 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3OYR
 
 | | Crystal structure of polyprenyl synthase from Caulobacter crescentus CB15 complexed with calcium and isoprenyl diphosphate | | Descriptor: | CALCIUM ION, DIMETHYLALLYL DIPHOSPHATE, PYROPHOSPHATE 2-, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Poulter, C.D, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-23 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3LOM
 
 | | CRYSTAL STRUCTURE OF GERANYLTRANSFERASE FROM Legionella pneumophila | | Descriptor: | Geranyltranstransferase, PHOSPHATE ION | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-04 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3LVS
 
 | | Crystal structure of farnesyl diphosphate synthase from rhodobacter capsulatus sb1003 | | Descriptor: | FARNESYL DIPHOSPHATE SYNTHASE, GLYCEROL, PHOSPHATE ION | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-22 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3MZV
 
 | | Crystal structure of a decaprenyl diphosphate synthase from Rhodobacter capsulatus | | Descriptor: | Decaprenyl diphosphate synthase | | Authors: | Quartararo, C.E, Patskovsky, Y, Bonanno, J.B, Rutter, M, Bain, K.T, Chang, S, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-13 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3NO8
 
 | | Crystal structure of the PHR domain from human BTBD2 Protein | | Descriptor: | BTB/POZ domain-containing protein 2, GLYCEROL, SULFATE ION | | Authors: | Sampathkumar, P, Miller, S, Rutter, M, Bain, K, Gheyi, T, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-24 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the PHR domain from human BTBD2 Protein
To be Published
|
|
3H14
 
 | | Crystal structure of a putative aminotransferase from Silicibacter pomeroyi | | Descriptor: | Aminotransferase, classes I and II, GLYCEROL | | Authors: | Sampathkumar, P, Atwell, S, Wasserman, S, Miller, S, Bain, K, Rutter, M, Tarun, G, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-10 | | Release date: | 2009-05-05 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative aminotransferase from Silicibacter pomeroyi
TO BE PUBLISHED
|
|
1F9E
 
 | | CASPASE-8 SPECIFICITY PROBED AT SUBSITE S4: CRYSTAL STRUCTURE OF THE CASPASE-8-Z-DEVD-CHO | | Descriptor: | (PHQ)DEVD, Caspase-8 subunit p10, Caspase-8 subunit p18 | | Authors: | Blanchard, H, Donepudi, M, Tschopp, M, Kodandapani, L, Wu, J.C, Grutter, M.G. | | Deposit date: | 2000-07-10 | | Release date: | 2001-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Caspase-8 specificity probed at subsite S(4): crystal structure of the caspase-8-Z-DEVD-cho complex.
J.Mol.Biol., 302, 2000
|
|
1TZE
 
 | |
3SQB
 
 | | Structure of the major type 1 pilus subunit FimA bound to the FimC chaperone | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Chaperone protein fimC, ... | | Authors: | Scharer, M.A, Eidam, O, Grutter, M.G, Glockshuber, R, Capitani, G. | | Deposit date: | 2011-07-05 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Quality control of disulfide bond formation in pilus subunits by the chaperone FimC.
Nat.Chem.Biol., 8, 2012
|
|
1ZE3
 
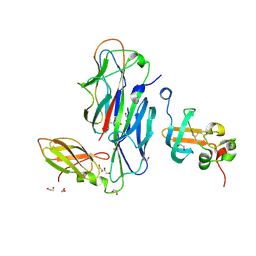 | | Crystal Structure of the Ternary Complex of FIMD (N-Terminal Domain) with FIMC and the Pilin Domain of FIMH | | Descriptor: | 1,2-ETHANEDIOL, Chaperone protein fimC, FimH protein, ... | | Authors: | Nishiyama, M, Horst, R, Eidam, O, Herrmann, T, Ignatov, O, Vetsch, M, Bettendorff, P, Jelesarov, I, Grutter, M.G, Wuthrich, K, Glockshuber, R, Capitani, G. | | Deposit date: | 2005-04-17 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD.
Embo J., 24, 2005
|
|
1ZDX
 
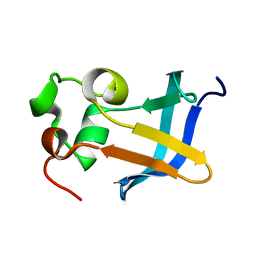 | | Solution Structure of the type 1 pilus assembly platform FimD(25-125) | | Descriptor: | Outer membrane usher protein fimD | | Authors: | Nishiyama, M, Horst, R, Herrmann, T, Vetsch, M, Bettendorff, P, Ignatov, O, Grutter, M, Wuthrich, K, Glockshuber, R, Capitani, G. | | Deposit date: | 2005-04-15 | | Release date: | 2005-06-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD.
Embo J., 24, 2005
|
|
3TJE
 
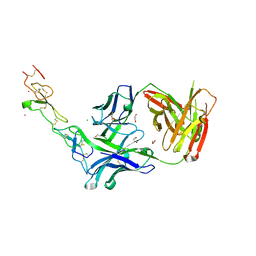 | | Crystal structure of Fas receptor extracellular domain in complex with Fab E09 | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Zuger, S, Stirnimann, C, Briand, C, Grutter, M.G. | | Deposit date: | 2011-08-24 | | Release date: | 2012-05-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A series of Fas receptor agonist antibodies that demonstrate an inverse correlation between affinity and potency.
Cell Death Differ., 19, 2012
|
|
1ZDV
 
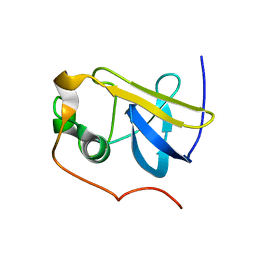 | | Solution Structure of the type 1 pilus assembly platform FimD(25-139) | | Descriptor: | Outer membrane usher protein fimD | | Authors: | Nishiyama, M, Horst, R, Herrmann, T, Vetsch, M, Bettendorff, P, Ignatov, O, Grutter, M, Wuthrich, K, Glockshuber, R, Capitani, G. | | Deposit date: | 2005-04-15 | | Release date: | 2005-06-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD.
Embo J., 24, 2005
|
|
3THM
 
 | | Crystal structure of Fas receptor extracellular domain in complex with Fab EP6b_B01 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Fab EP6b_B01, ... | | Authors: | Zuger, S, Stirnimann, C, Briand, C, Grutter, M.G. | | Deposit date: | 2011-08-19 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A series of Fas receptor agonist antibodies that demonstrate an inverse correlation between affinity and potency.
Cell Death Differ., 19, 2012
|
|
4DB6
 
 | | Designed Armadillo repeat protein (YIIIM3AII) | | Descriptor: | Armadillo repeat protein | | Authors: | Madhurantakam, C, Varadamsetty, G, Grutter, M.G, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2012-01-13 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based optimization of designed Armadillo-repeat proteins.
Protein Sci., 21, 2012
|
|
4DB8
 
 | | Designed Armadillo-repeat Protein | | Descriptor: | Armadillo-repeat Protein, MAGNESIUM ION | | Authors: | Madhurantakam, C, Varadamsetty, G, Grutter, M.G, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2012-01-13 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based optimization of designed Armadillo-repeat proteins.
Protein Sci., 21, 2012
|
|
1YNU
 
 | | Crystal structure of apple ACC synthase in complex with L-vinylglycine | | Descriptor: | 1-aminocyclopropane-1-carboxylate synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[O-PHOSPHONOPYRIDOXYL]-AMINO- BUTYRIC ACID, ... | | Authors: | Capitani, G, Tschopp, M, Eliot, A.C, Kirsch, J.F, Grutter, M.G. | | Deposit date: | 2005-01-25 | | Release date: | 2005-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of ACC synthase inactivated by the mechanism-based inhibitor L-vinylglycine.
Febs Lett., 579, 2005
|
|
4DBA
 
 | | Designed Armadillo repeat protein (YIIM3AII) | | Descriptor: | Designed Armadillo repeat protein, YIIM3AII, GLYCEROL | | Authors: | Madhurantakam, C, Varadamsetty, G, Grutter, M.G, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2012-01-13 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based optimization of designed Armadillo-repeat proteins.
Protein Sci., 21, 2012
|
|
4DB9
 
 | | Designed Armadillo repeat protein (YIIIM3AIII) | | Descriptor: | Armadillo repeat protein, YIIIM3AIII | | Authors: | Madhurantakam, C, Varadamsetty, G, Grutter, M.G, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2012-01-13 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based optimization of designed Armadillo-repeat proteins.
Protein Sci., 21, 2012
|
|
1J0X
 
 | | Crystal structure of the rabbit muscle glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Cowan-Jacob, S.W, Kaufmann, M, Anselmo, A.N, Stark, W, Grutter, M.G. | | Deposit date: | 2002-11-25 | | Release date: | 2003-12-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of rabbit-muscle glyceraldehyde-3-phosphate dehydrogenase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1JPF
 
 | | Crystal Structure Of The LCMV Peptidic Epitope Gp276 In Complex With The Murine Class I Mhc Molecule H-2Db | | Descriptor: | BETA-2-MICROGLOBULIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-B ALPHA CHAIN, ... | | Authors: | Ciatto, C, Tissot, A.C, Tschopp, M, Capitani, G, Pecorari, F, Pluckthun, A, Grutter, M.G. | | Deposit date: | 2001-08-02 | | Release date: | 2001-10-24 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Zooming in on the hydrophobic ridge of H-2D(b): implications for the conformational variability of bound peptides.
J.Mol.Biol., 312, 2001
|
|
3PIU
 
 | |
1JPG
 
 | | Crystal Structure Of The LCMV Peptidic Epitope Np396 In Complex With The Murine Class I Mhc Molecule H-2Db | | Descriptor: | BETA-2-MICROGLOBULIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-B ALPHA CHAIN, ... | | Authors: | Ciatto, C, Tissot, A.C, Tschopp, M, Capitani, G, Pecorari, F, Pluckthun, A, Grutter, M.G. | | Deposit date: | 2001-08-02 | | Release date: | 2001-10-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Zooming in on the hydrophobic ridge of H-2D(b): implications for the conformational variability of bound peptides.
J.Mol.Biol., 312, 2001
|
|
2V12
 
 | | Crystal Structure of Renin with Inhibitor 8 | | Descriptor: | N-[(2S,4S,5S,7R)-4-AMINO-8-(BUTYLAMINO)-5-HYDROXY-7-METHYL-2-(1-METHYLETHYL)-8-OXOOCTYL]-2-(3-METHOXYPROPOXY)BENZAMIDE, RENIN | | Authors: | Rahuel, J, Rasetti, V, Maibaum, J, Rueger, H, Goschke, R, Cohen, N.C, Stutz, S, Cumin, F, Fuhrer, W, Wood, J.M, Grutter, M.G. | | Deposit date: | 2007-05-21 | | Release date: | 2007-07-03 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-Based Drug Design: The Discovery of Novel Nonpeptide Orally Active Inhibitors of Human Renin
Chem.Biol., 7, 2000
|
|
