8QSR
 
 | |
8QST
 
 | |
9HNP
 
 | |
9FMD
 
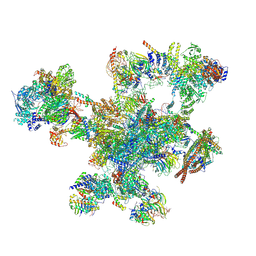 | | Integrative model of the human post-catalytic spliceosome (P-complex) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Rothe, P, Plaschka, C, Vorlaender, M.K. | | Deposit date: | 2024-06-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism for the initiation of spliceosome disassembly.
Nature, 632, 2024
|
|
8RO2
 
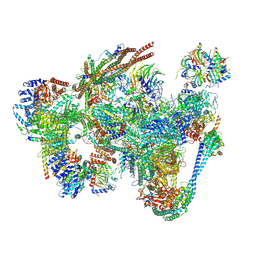 | | Integrative Structure of the human intron lariat Spliceosome (ILS'') | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ATP-dependent RNA helicase DHX15, CWF19-like protein 1, ... | | Authors: | Rothe, P, Vorlaender, M.K, Plaschka, C. | | Deposit date: | 2024-01-11 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism for the initiation of spliceosome disassembly.
Nature, 632, 2024
|
|
8RO0
 
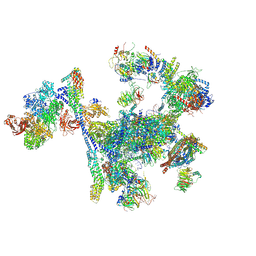 | | Structure of the C. elegans Intron Lariat Spliceosome primed for disassembly (ILS') | | Descriptor: | Cell division cycle 5-like protein, Coiled-coil domain-containing protein 12, GCF C-terminal domain-containing protein, ... | | Authors: | Vorlaender, M.K, Rothe, P, Plaschka, C. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-07 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism for the initiation of spliceosome disassembly.
Nature, 632, 2024
|
|
8RO1
 
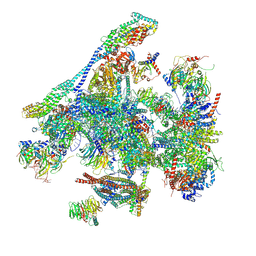 | | Structure of the C. elegans Intron Lariat Spliceosome double-primed for disassembly (ILS'') | | Descriptor: | CWF19-like protein 1 homolog, CWF19-like protein 2 homolog, Cell division cycle 5-like protein, ... | | Authors: | Vorlaender, M.K, Rothe, P, Plaschka, C. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism for the initiation of spliceosome disassembly.
Nature, 632, 2024
|
|
8C8Q
 
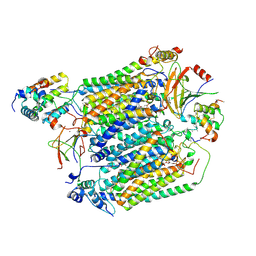 | | Cytochrome c oxidase from Schizosaccharomyces pombe | | Descriptor: | CALCIUM ION, COPPER (II) ION, Cytochrome c oxidase polypeptide 5, ... | | Authors: | Moe, A, Adelroth, P, Brzezinski, P, Nasvik Ojemyr, L. | | Deposit date: | 2023-01-20 | | Release date: | 2023-03-01 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Cryo-EM structure and function of S. pombe complex IV with bound respiratory supercomplex factor.
Commun Chem, 6, 2023
|
|
5CTV
 
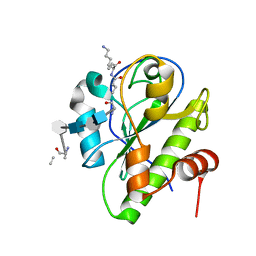 | | Catalytic domain of LytA, the major autolysin of Streptococcus pneumoniae, (C60A, H133A, C136A mutant) complexed with peptidoglycan fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, Autolysin, fragment of peptidoglycan | | Authors: | Achour, A, Sandalova, T, Mellroth, P. | | Deposit date: | 2015-07-24 | | Release date: | 2016-06-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The crystal structure of the major pneumococcal autolysin LytA in complex with a large peptidoglycan fragment reveals the pivotal role of glycans for lytic activity.
Mol.Microbiol., 101, 2016
|
|
4IWT
 
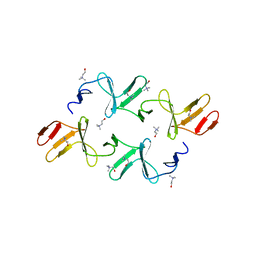 | |
6LUL
 
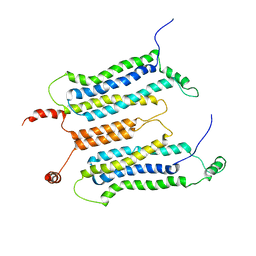 | | NMR structure and dynamics studies of yeast respiratory super-complex factor 2 in micelles | | Descriptor: | Respiratory supercomplex factor 2, mitochondrial | | Authors: | Zhou, S, Pontus, P, Peter, B, Maler, L, Adelroth, P. | | Deposit date: | 2020-01-29 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Dynamics Studies of Yeast Respiratory Supercomplex Factor 2.
Structure, 29, 2021
|
|
2NX9
 
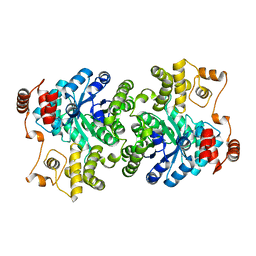 | |
5A7R
 
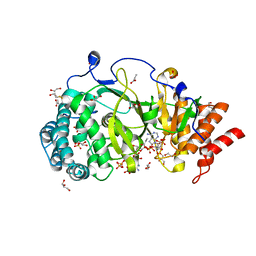 | | Human poly(ADP-ribose) glycohydrolase in complex with synthetic dimeric ADP-ribose | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, POLY(ADP-RIBOSE) GLYCOHYDROLASE, ... | | Authors: | Lambrecht, M.J, Brichacek, M, Barkauskaite, E, Ariza, A, Ahel, I, Hergenrother, P.J. | | Deposit date: | 2015-07-09 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Synthesis of Dimeric Adp-Ribose and its Structure with Human Poly(Adp-Ribose) Glycohydrolase.
J.Am.Chem.Soc., 137, 2015
|
|
6ETJ
 
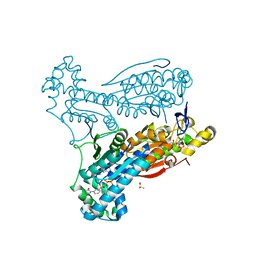 | | HUMAN PFKFB3 IN COMPLEX WITH KAN0438241 | | Descriptor: | 4-[[3-(5-fluoranyl-2-oxidanyl-phenyl)phenyl]sulfonylamino]-2-oxidanyl-benzoic acid, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | Authors: | Gustafsson, N.M.S, Lundback, T, Farnegardh, K, Groth, P, Wiitta, E, Jonsson, M, Hallberg, K, Pennisi, R, Huguet Ninou, A, Martinsson, J, Norstrom, C, Schultz, J, Andersson, M, Markova, N, Marttila, P, Norin, M, Olin, T, Helleday, T. | | Deposit date: | 2017-10-26 | | Release date: | 2018-11-07 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Targeting PFKFB3 radiosensitizes cancer cells and suppresses homologous recombination.
Nat Commun, 9, 2018
|
|
1YCE
 
 | | Structure of the rotor ring of F-type Na+-ATPase from Ilyobacter tartaricus | | Descriptor: | NONAN-1-OL, SODIUM ION, subunit c | | Authors: | Meier, T, Polzer, P, Diederichs, K, Welte, W, Dimroth, P. | | Deposit date: | 2004-12-22 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the rotor ring of F-Type Na+-ATPase from Ilyobacter tartaricus.
Science, 308, 2005
|
|
1ISK
 
 | | 3-OXO-DELTA5-STEROID ISOMERASE, NMR, 20 STRUCTURES | | Descriptor: | 3-OXO-DELTA5-STEROID ISOMERASE | | Authors: | Wu, Z.R, Ebrahimian, S, Zawrotny, M.E, Thornburg, L.D, Perez-Alvarado, G.C, Brothers, P, Pollack, R.M, Summers, M.F. | | Deposit date: | 1997-03-12 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of 3-oxo-delta5-steroid isomerase.
Science, 276, 1997
|
|
2QE7
 
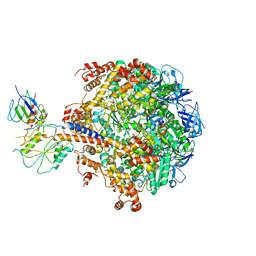 | | Crystal structure of the f1-atpase from the thermoalkaliphilic bacterium bacillus sp. ta2.a1 | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit epsilon, ... | | Authors: | Stocker, A, Keis, S, Vonck, J, Cook, G.M, Dimroth, P. | | Deposit date: | 2007-06-25 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | The Structural Basis for Unidirectional Rotation of Thermoalkaliphilic F(1)-ATPase.
Structure, 15, 2007
|
|
