4KVK
 
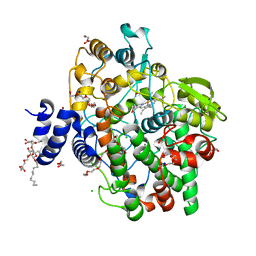 | | Crystal structure of Oryza sativa fatty acid alpha-dioxygenase | | Descriptor: | CALCIUM ION, CHLORIDE ION, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Zhu, G, Koszelak-Rosenblum, M, Malkowski, M.G. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of alpha-dioxygenase from Oryza sativa: Insights into substrate binding and activation by hydrogen peroxide.
Protein Sci., 22, 2013
|
|
8JJE
 
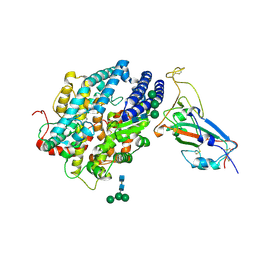 | | RBD of SARS-CoV2 spike protein with ACE2 decoy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Kishikawa, J, Hirose, M, Kato, T, Okamoto, T. | | Deposit date: | 2023-05-30 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An inhaled ACE2 decoy confers protection against SARS-CoV-2 infection in preclinical models.
Sci Transl Med, 15, 2023
|
|
4KVJ
 
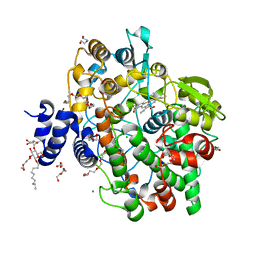 | | Crystal structure of Oryza sativa fatty acid alpha-dioxygenase with hydrogen peroxide | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Zhu, G, Koszelak-Rosenblum, M, Malkowski, M.G. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structures of alpha-dioxygenase from Oryza sativa: Insights into substrate binding and activation by hydrogen peroxide.
Protein Sci., 22, 2013
|
|
4KVL
 
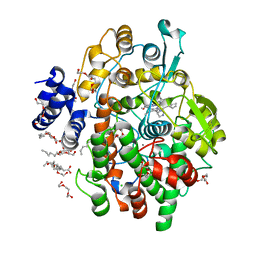 | | Crystal structure of Oryza sativa fatty acid alpha-dioxygenase Y379F with palmitic acid | | Descriptor: | CALCIUM ION, CHLORIDE ION, Fatty acid alpha-oxidase, ... | | Authors: | Zhu, G, Koszelak-Rosenblum, M, Malkowski, M.G. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structures of alpha-dioxygenase from Oryza sativa: Insights into substrate binding and activation by hydrogen peroxide.
Protein Sci., 22, 2013
|
|
1BL1
 
 | | PTH RECEPTOR N-TERMINUS FRAGMENT, NMR, 1 STRUCTURE | | Descriptor: | PARATHYROID HORMONE RECEPTOR | | Authors: | Pellegrini, M, Bisello, A, Rosenblatt, M, Chorev, M, Mierke, D.F. | | Deposit date: | 1998-07-22 | | Release date: | 1999-03-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Binding domain of human parathyroid hormone receptor: from conformation to function.
Biochemistry, 37, 1998
|
|
7DZY
 
 | | Spike protein from SARS-CoV2 with Fab fragment of enhancing antibody 2490 | | Descriptor: | Fab Heavy chain of enhancing antibody 2490, Fab light chain of enhancing antibody 2490, Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
7DZX
 
 | | Spike protein from SARS-CoV2 with Fab fragment of enhancing antibody 8D2 | | Descriptor: | Fab Heavy chain of enhancing antibody, Fab light chain of enhancing antibody, Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
7DZW
 
 | | Apo spike protein from SARS-CoV2 | | Descriptor: | Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
1CEE
 
 | | SOLUTION STRUCTURE OF CDC42 IN COMPLEX WITH THE GTPASE BINDING DOMAIN OF WASP | | Descriptor: | GTP-BINDING RHO-LIKE PROTEIN, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Abdul-Manan, N, Aghazadeh, B, Liu, G.A, Majumdar, A, Ouerfelli, O, Rosen, M.K. | | Deposit date: | 1999-03-08 | | Release date: | 1999-06-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of Cdc42 in complex with the GTPase-binding domain of the 'Wiskott-Aldrich syndrome' protein.
Nature, 399, 1999
|
|
7D68
 
 | | Cryo-EM structure of the human glucagon-like peptide-2 receptor-Gs protein complex | | Descriptor: | Glucagon-like peptide 2 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, W, Chen, L, Zhou, Q, Zhao, L, Zhang, H, Cong, Z, Shen, D, Zhao, F, Zhou, F, Cai, X, Chen, Y, Zhou, Y, Gadgaard, S, van der Velden, W.J, Zhao, S, Jiang, Y, Rosenkilde, M.M, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A unique hormonal recognition feature of the human glucagon-like peptide-2 receptor.
Cell Res., 30, 2020
|
|
6GK8
 
 | | Crystal structure of anti-tau antibody dmCBTAU-28.1, double mutant (S32R, E35K) of CBTAU-28.1, in complex with Tau peptide A7731 (residues 52-71) | | Descriptor: | CHLORIDE ION, HUMAN FAB ANTIBODY FRAGMENT OF CBTAU-28.1(S32R;E35K), TAU PEPTIDE A7731 (RESIDUES 52-71) | | Authors: | Steinbacher, S, Mrosek, M, Juraszek, J. | | Deposit date: | 2018-05-18 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A common antigenic motif recognized by naturally occurring human VH5-51/VL4-1 anti-tau antibodies with distinct functionalities.
Acta Neuropathol Commun, 6, 2018
|
|
6GK7
 
 | | Crystal structure of anti-tau antibody dmCBTAU-27.1, double mutant (S31Y, T100I) of CBTAU-27.1, in complex with Tau peptide A8119B (residues 299-318) | | Descriptor: | CHLORIDE ION, HUMAN FAB ANTIBODY FRAGMENT OF CBTAU-27.1(S31Y,T100I), HUMAN TAU PEPTIDE A8119 RESIDUES 299-318 | | Authors: | Steinbacher, S, Mrosek, M, Juraszek, J. | | Deposit date: | 2018-05-18 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A common antigenic motif recognized by naturally occurring human VH5-51/VL4-1 anti-tau antibodies with distinct functionalities.
Acta Neuropathol Commun, 6, 2018
|
|
5LVX
 
 | | Crystal structure of glucocerebrosidase with an inhibitory quinazoline modulator | | Descriptor: | 11-[(2~{R})-2-[(2-pyridin-3-ylquinazolin-4-yl)amino]-2,3-dihydro-1~{H}-inden-5-yl]undec-10-ynoic acid, 11-[(2~{S})-2-[(2-pyridin-3-ylquinazolin-4-yl)amino]-2,3-dihydro-1~{H}-inden-5-yl]undec-10-ynoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zheng, J, Chen, L, Skinner, O.S, Lansbury, P, Skerlj, R, Mrosek, M, Heunisch, U, Krapp, S, Weigand, S, Charrow, J, Schwake, M, Kelleher, N.L, Silverman, R.B, Krainc, D. | | Deposit date: | 2016-09-14 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | beta-Glucocerebrosidase Modulators Promote Dimerization of beta-Glucocerebrosidase and Reveal an Allosteric Binding Site.
J. Am. Chem. Soc., 140, 2018
|
|
6WNL
 
 | | human Artemis/SNM1C catalytic domain, crystal form 2 | | Descriptor: | Protein artemis, ZINC ION | | Authors: | Karim, F, Liu, S, Laciak, A.R, Volk, L, Rosenblum, M, Curtis, R, Huang, N, Carr, G, Zhu, G. | | Deposit date: | 2020-04-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural analysis of the catalytic domain of Artemis endonuclease/SNM1C reveals distinct structural features.
J.Biol.Chem., 295, 2020
|
|
8R6Y
 
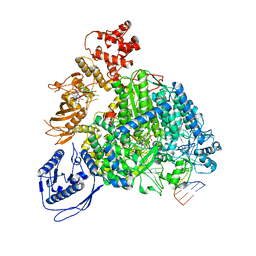 | | Structure of the SFTSV L protein stalled in a transcription-specific early elongation state with bound capped RNA [TRANSCRIPTION-EARLY-ELONGATION] | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, MAGNESIUM ION, RNA (5'-R(*(M7G)*AP*AP*A)-3'), ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Busch, C, Milewski, M, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2023-11-23 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural snapshots of phenuivirus cap-snatching and transcription.
Nucleic Acids Res., 52, 2024
|
|
8R6W
 
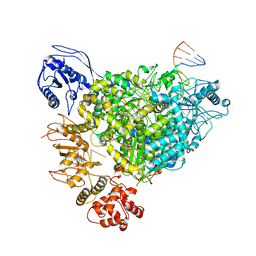 | | Structure of the SFTSV L protein in a transcription-priming state with bound capped RNA [TRANSCRIPTION-PRIMING] | | Descriptor: | RNA (5'-R(*(M7G)*AP*AP*A)-3'), RNA (5'-R(*AP*CP*AP*C)-3'), RNA (5'-R(*AP*CP*AP*CP*AP*GP*AP*GP*AP*CP*GP*CP*CP*CP*AP*G)-3'), ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Busch, C, Milewski, M, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2023-11-23 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural snapshots of phenuivirus cap-snatching and transcription.
Nucleic Acids Res., 52, 2024
|
|
8R6U
 
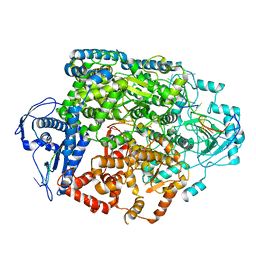 | | Structure of the SFTSV L protein in a transcription-priming state without capped RNA [TRANSCRIPTION-PRIMING (in vitro)] | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*CP*UP*GP*GP*GP*CP*GP*GP*UP*CP*UP*UP*UP*GP*UP*GP*U)-3'), RNA primer, ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Busch, C, Milewski, M, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2023-11-23 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural snapshots of phenuivirus cap-snatching and transcription.
Nucleic Acids Res., 52, 2024
|
|
6T5J
 
 | | Structure of NUDT15 in complex with inhibitor TH1760 | | Descriptor: | 6-[4-(1~{H}-indol-5-ylcarbonyl)piperazin-1-yl]sulfonyl-3~{H}-1,3-benzoxazol-2-one, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Carter, M, Rehling, D, Desroses, M, Zhang, S.M, Hagenkort, A, Valerie, N.C.K, Helleday, T, Stenmark, P. | | Deposit date: | 2019-10-16 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a chemical probe against NUDT15.
Nat.Chem.Biol., 16, 2020
|
|
6S4A
 
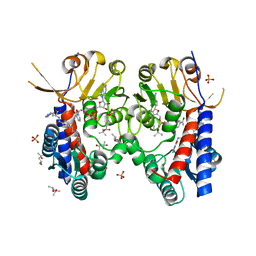 | | Structure of human MTHFD2 in complex with TH9028 | | Descriptor: | (2~{S})-2-[[5-[[2,4-bis(azanyl)-6-oxidanylidene-5~{H}-pyrimidin-5-yl]carbamoylamino]pyridin-2-yl]carbonylamino]-4-(1~{H}-1,2,3,4-tetrazol-5-yl)butanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, ... | | Authors: | Gustafsson, R, Scaletti, E.R, Bonagas, N, Gustafsson, N.M, Henriksson, M, Abdurakhmanov, E, Andersson, Y, Bengtsson, C, Borhade, S, Desroses, M, Farnegardh, K, Garg, N, Gokturk, C, Haraldsson, M, Iliev, P, Jarvius, M, Jemth, A.S, Kalderen, C, Karsten, S, Klingegard, F, Koolmeister, T, Martens, U, Llona-Minguez, S, Loseva, O, Marttila, P, Michel, M, Moulson, R, Nordstrom, H, Paulin, C, Pham, T, Pudelko, L, Rasti, A, Roos, A.K, Sarno, A, Sandberg, L, Scobie, M, Sjoberg, B, Svensson, R, Unterlass, J.E, Vallin, K, Vo, D, Wiita, E, Warpman-Berglund, U, Homan, E.J, Helleday, T, Stenmark, P. | | Deposit date: | 2019-06-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress.
Nat Cancer, 3, 2022
|
|
6S4F
 
 | | Structure of human MTHFD2 in complex with TH9619 | | Descriptor: | (E,4S)-4-[[5-[2-[2,6-bis(azanyl)-4-oxidanylidene-1H-pyrimidin-5-yl]ethanoylamino]-3-fluoranyl-pyridin-2-yl]carbonylamino]pent-2-enedioic acid, ADENOSINE-5'-DIPHOSPHATE, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, ... | | Authors: | Scaletti, E.R, Gustafsson, R, Bonagas, N, Gustafsson, N.M, Henriksson, M, Abdurakhmanov, E, Andersson, Y, Bengtsson, C, Borhade, S, Desroses, M, Farnegardh, K, Garg, N, Gokturk, C, Haraldsson, M, Iliev, P, Jarvius, M, Jemth, A.S, Kalderen, C, Karsten, S, Klingegard, F, Koolmeister, T, Martens, U, Llona-Minguez, S, Loseva, O, Marttila, P, Michel, M, Moulson, R, Nordstrom, H, Paulin, C, Pham, T, Pudelko, L, Rasti, A, Roos, A.K, Sarno, A, Sandberg, L, Scobie, M, Sjoberg, B, Svensson, R, Unterlass, J.E, Vallin, K, Vo, D, Wiita, E, Warpman-Berglund, U, Homan, E.J, Helleday, T, Stenmark, P. | | Deposit date: | 2019-06-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress.
Nat Cancer, 3, 2022
|
|
6S4E
 
 | | Structure of human MTHFD2 in complex with TH7299 | | Descriptor: | (2S)-2-[[4-[[2,4-bis(azanyl)-6-oxidanylidene-1H-pyrimidin-5-yl]carbamoylamino]phenyl]carbonylamino]pentanedioic acid, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, ... | | Authors: | Gustafsson, R, Scaletti, E.R, Bonagas, N, Gustafsson, N.M, Henriksson, M, Abdurakhmanov, E, Andersson, Y, Bengtsson, C, Borhade, S, Desroses, M, Farnegardh, K, Garg, N, Gokturk, C, Haraldsson, M, Iliev, P, Jarvius, M, Jemth, A.S, Kalderen, C, Karsten, S, Klingegard, F, Koolmeister, T, Martens, U, Llona-Minguez, S, Loseva, O, Marttila, P, Michel, M, Moulson, R, Nordstrom, H, Paulin, C, Pham, T, Pudelko, L, Rasti, A, Roos, A.K, Sarno, A, Sandberg, L, Scobie, M, Sjoberg, B, Svensson, R, Unterlass, J.E, Vallin, K, Vo, D, Wiita, E, Warpman-Berglund, U, Homan, E.J, Helleday, T, Stenmark, P. | | Deposit date: | 2019-06-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress.
Nat Cancer, 3, 2022
|
|
6SIU
 
 | | Crystal structure of IbpAFic2 covalently tethered to Cdc42 | | Descriptor: | Cell division control protein 42 homolog, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gulen, B, Roselin, M, Albers, M, Hedberg, C, Itzen, A, Pogenberg, V. | | Deposit date: | 2019-08-12 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Identification of targets of AMPylating Fic enzymes by co-substrate-mediated covalent capture.
Nat.Chem., 12, 2020
|
|
6LXV
 
 | | Cryo-EM structure of phosphoketolase from Bifidobacterium longum | | Descriptor: | CALCIUM ION, Phosphoketolase, THIAMINE DIPHOSPHATE | | Authors: | Nakata, K, Miyazaki, N, Yamaguchi, H, Hirose, M, Miyano, H, Mizukoshi, T, Kashiwagi, T, Iwasaki, K. | | Deposit date: | 2020-02-12 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | High-resolution structure of phosphoketolase from Bifidobacterium longum determined by cryo-EM single-particle analysis.
J.Struct.Biol., 214, 2022
|
|
6XYA
 
 | | Cap-binding domain of SFTSV L protein | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, RNA-dependent RNA polymerase, SODIUM ION | | Authors: | Gogrefe, N, Guenther, S, Rosenthal, M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and functional characterization of the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 48, 2020
|
|
8Z4D
 
 | | Structure of the S-ring region of the Vibrio flagellar MS-ring protein FliF with 34-fold symmetry applied | | Descriptor: | Flagellar M-ring protein,Flagellar motor switch protein FliG | | Authors: | Takekawa, N, Nishikino, T, Kishikawa, J, Hirose, M, Kato, T, Imada, K, Homma, M. | | Deposit date: | 2024-04-17 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural analysis of S-ring composed of FliFG fusion proteins in marine Vibrio polar flagellar motors
To Be Published
|
|
