7ZBU
 
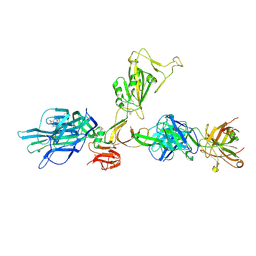 | | CryoEM structure of SARS-CoV-2 spike monomer in complex with neutralising antibody P008_60 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-2-[[5-[(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, P008_60 antibody, ... | | Authors: | Rosa, A, Pye, V.E, Cronin, N, Cherepanov, P. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | A neutralizing epitope on the SD1 domain of SARS-CoV-2 spike targeted following infection and vaccination.
Cell Rep, 40, 2022
|
|
7NT9
 
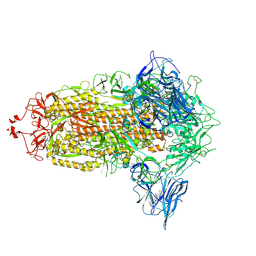 | | Trimeric SARS-CoV-2 spike ectodomain in complex with biliverdin (closed conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Rosa, A, Pye, V.E, Nans, A, Cherepanov, P. | | Deposit date: | 2021-03-09 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity.
Sci Adv, 7, 2021
|
|
7NTA
 
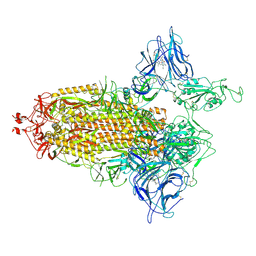 | | Trimeric SARS-CoV-2 spike ectodomain in complex with biliverdin (one RBD erect) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Rosa, A, Pye, V.E, Nans, A, Cherepanov, P. | | Deposit date: | 2021-03-09 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity.
Sci Adv, 7, 2021
|
|
7NTC
 
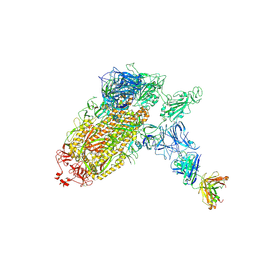 | | Trimeric SARS-CoV-2 spike ectodomain bound to P008_056 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Rosa, A, Pye, V.E, Nans, A, Cherepanov, P. | | Deposit date: | 2021-03-09 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity.
Sci Adv, 7, 2021
|
|
6PPW
 
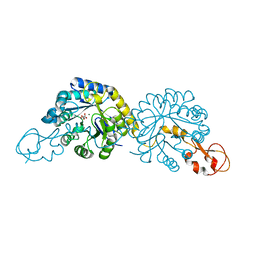 | | Crystal structure of NeuB, an N-acetylneuraminate synthase from Neisseria meningitidis, in complex with magnesium and malate | | Descriptor: | D-MALATE, MAGNESIUM ION, N-acetylneuraminate synthase | | Authors: | Rosanally, A.Z, Junop, M.S, Berti, P.J. | | Deposit date: | 2019-07-08 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | NeuNAc Oxime: A Slow-Binding and Effectively Irreversible Inhibitor of the Sialic Acid Synthase NeuB.
Biochemistry, 58, 2019
|
|
6PPX
 
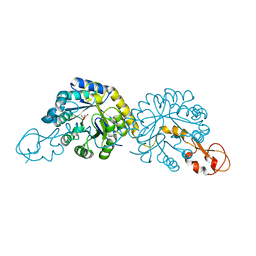 | |
6PPY
 
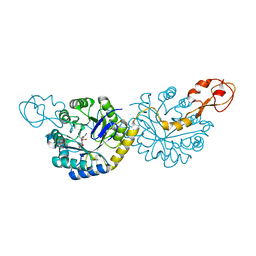 | | Crystal structure of NeuNAc oxime complexed with NeuB, an N-acetylneuraminate synthase from Neisseria meningitidis | | Descriptor: | (2E,4S,5R,6R,7S,8R)-5-(acetylamino)-4,6,7,8,9-pentahydroxy-2-(hydroxyimino)nonanoic acid (non-preferred name), N-acetylneuraminate synthase | | Authors: | Rosanally, A.Z, Junop, M.J, Berti, P.J. | | Deposit date: | 2019-07-08 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | NeuNAc Oxime: A Slow-Binding and Effectively Irreversible Inhibitor of the Sialic Acid Synthase NeuB.
Biochemistry, 58, 2019
|
|
7B62
 
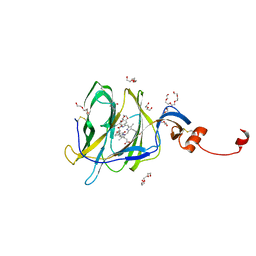 | | Crystal structure of SARS-CoV-2 spike protein N-terminal domain in complex with biliverdin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pye, V.E, Rosa, A, Roustan, C, Cherepanov, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity.
Sci Adv, 7, 2021
|
|
3QAC
 
 | | Structure of amaranth 11S proglobulin seed storage protein from Amaranthus hypochondriacus L. | | Descriptor: | 11S globulin seed storage protein | | Authors: | Tandang-Silvas, M.R, Carrazco-Pena, L, Barba de la Rosa, A.P, Osuna-Castro, J.A, Utsumi, S, Mikami, B, Maruyama, N. | | Deposit date: | 2011-01-10 | | Release date: | 2012-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.275 Å) | | Cite: | Structure of amaranth 11S proglobulin, a major seed storage protein from Amaranthus hypochondriacus L.
To be Published
|
|
1NOF
 
 | | THE FIRST CRYSTALLOGRAPHIC STRUCTURE OF A XYLANASE FROM GLYCOSYL HYDROLASE FAMILY 5: IMPLICATIONS FOR CATALYSIS | | Descriptor: | ACETATE ION, xylanase | | Authors: | Larson, S.B, Day, J, McPherson, A, Barba De La Rosa, A.P, Keen, N.T. | | Deposit date: | 2003-01-16 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | First crystallographic structure of a xylanase from glycoside hydrolase family 5: implications for catalysis.
Biochemistry, 42, 2003
|
|
4IYD
 
 | |
4IYF
 
 | |
1Y3K
 
 | | Solution structure of the apo form of the fifth domain of Menkes protein | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Chasapis, C.T, Ciofi-Baffoni, S, Hadjiliadis, N, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-11-25 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An NMR study of the interaction between the human copper(I) chaperone and the second and fifth metal-binding domains of the Menkes protein
Febs J., 272, 2005
|
|
5T7L
 
 | | Pt(II)-mediated copper-dependent interactions between ATOX1 and MNK1 | | Descriptor: | COPPER (II) ION, Copper transport protein ATOX1, Copper-transporting ATPase 1, ... | | Authors: | Caliandro, R, Mirabelli, V, Caliandro, R, Rosato, A, Lasorsa, A, Galliani, A, Arnesano, F, Natile, G. | | Deposit date: | 2016-09-05 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Mechanistic and Structural Basis for Inhibition of Copper Trafficking by Platinum Anticancer Drugs.
J.Am.Chem.Soc., 141, 2019
|
|
1Y3J
 
 | | Solution structure of the copper(I) form of the fifth domain of Menkes protein | | Descriptor: | COPPER (II) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Chasapis, C.T, Ciofi-Baffoni, S, Hadjiliadis, N, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-11-25 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An NMR study of the interaction between the human copper(I) chaperone and the second and fifth metal-binding domains of the Menkes protein
Febs J., 272, 2005
|
|
1AQA
 
 | | SOLUTION STRUCTURE OF REDUCED MICROSOMAL RAT CYTOCHROME B5, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Banci, L, Bertini, I, Ferroni, F, Rosato, A. | | Deposit date: | 1997-07-28 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of reduced microsomal rat cytochrome b5.
Eur.J.Biochem., 249, 1997
|
|
1YJU
 
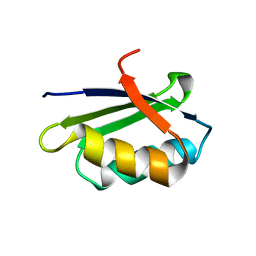 | | Solution structure of the apo form of the sixth soluble domain of Menkes protein | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Rosato, A, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-15 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An atomic-level investigation of the disease-causing A629P mutant of the Menkes protein, ATP7A
J.Mol.Biol., 352, 2005
|
|
1YJR
 
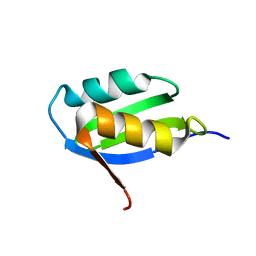 | | Solution structure of the apo form of the sixth soluble domain A69P mutant of Menkes protein | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Rosato, A, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-15 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An atomic-level investigation of the disease-causing A629P mutant of the Menkes protein, ATP7A
J.Mol.Biol., 352, 2005
|
|
1S6O
 
 | | Solution structure and backbone dynamics of the apo-form of the second metal-binding domain of the Menkes protein ATP7A | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Del Conte, R, D'Onofrio, M, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-01-26 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the Cu(I) and Apo Forms of the Second Metal-Binding Domain of the Menkes Protein ATP7A.
Biochemistry, 43, 2004
|
|
1BFY
 
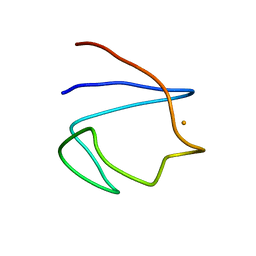 | | SOLUTION STRUCTURE OF REDUCED CLOSTRIDIUM PASTEURIANUM RUBREDOXIN, NMR, 20 STRUCTURES | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Bertini, I, Kurtz Junior, D.M, Eidsness, M.K, Liu, G, Luchinat, C, Rosato, A, Scott, R.A. | | Deposit date: | 1998-05-23 | | Release date: | 1999-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Reduced Clostridium Pasteurianum Rubredoxin
J.Biol.Inorg.Chem., 3, 1998
|
|
1YJT
 
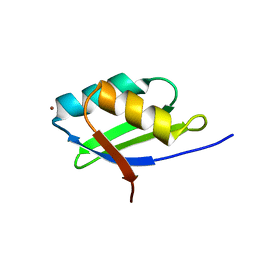 | | Solution structure of the Cu(I) form of the sixth soluble domain A69P mutant of Menkes protein | | Descriptor: | COPPER (I) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Rosato, A, Wang, S. | | Deposit date: | 2005-01-15 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An atomic-level investigation of the disease-causing A629P mutant of the Menkes protein, ATP7A
J.Mol.Biol., 352, 2005
|
|
1YJV
 
 | | Solution structure of the Cu(I) form of the sixth soluble domain of Menkes protein | | Descriptor: | COPPER (I) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Rosato, A, Wang, S. | | Deposit date: | 2005-01-15 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An atomic-level investigation of the disease-causing A629P mutant of the Menkes protein, ATP7A
J.Mol.Biol., 352, 2005
|
|
2G9O
 
 | | Solution structure of the apo form of the third metal-binding domain of ATP7A protein (Menkes Disease protein) | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, DellaMalva, N, Rosato, A, Herrmann, T, Wuthrich, K, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-07 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and intermolecular interactions of the third metal-binding domain of ATP7A, the Menkes disease protein.
J.Biol.Chem., 281, 2006
|
|
3CJK
 
 | | Crystal structure of the adduct HAH1-Cd(II)-MNK1. | | Descriptor: | CADMIUM ION, Copper transport protein ATOX1, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Calderone, V, Felli, I, Della-Malva, N, Pavelkova, A, Rosato, A. | | Deposit date: | 2008-03-13 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Copper(I)-mediated protein-protein interactions result from suboptimal interaction surfaces.
Biochem.J., 422, 2009
|
|
1AKK
 
 | | SOLUTION STRUCTURE OF OXIDIZED HORSE HEART CYTOCHROME C, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Banci, L, Bertini, I, Gray, H.B, Luchinat, C, Reddig, T, Rosato, A, Turano, P. | | Deposit date: | 1997-05-22 | | Release date: | 1997-09-17 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of oxidized horse heart cytochrome c.
Biochemistry, 36, 1997
|
|
