5JUP
 
 | | Saccharomyces cerevisiae 80S ribosome bound with elongation factor eEF2-GDP-sordarin and Taura Syndrome Virus IRES, Structure II (mid-rotated 40S subunit) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Abeyrathne, P, Koh, C.S, Grant, T, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-05 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome.
Elife, 5, 2016
|
|
5EPM
 
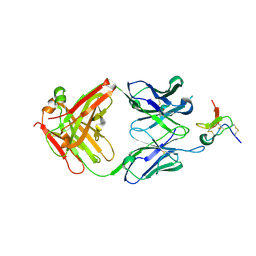 | |
1R65
 
 | | Crystal structure of ferrous soaked Ribonucleotide Reductase R2 subunit (wildtype) at pH 5 from E. coli | | Descriptor: | FE (II) ION, MERCURY (II) ION, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Voegtli, W.C, Sommerhalter, M, Saleh, L, Baldwin, J, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-10-14 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Variable coordination geometries at the diiron(II) active site of ribonucleotide reductase R2.
J.Am.Chem.Soc., 125, 2003
|
|
2B8E
 
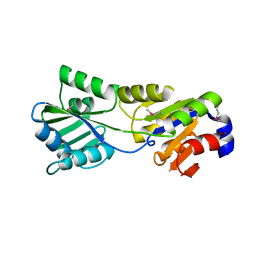 | |
1PM2
 
 | | CRYSTAL STRUCTURE OF MANGANESE SUBSTITUTED R2-D84E (D84E MUTANT OF THE R2 SUBUNIT OF E. COLI RIBONUCLEOTIDE REDUCTASE) | | Descriptor: | MANGANESE (II) ION, MERCURY (II) ION, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Voegtli, W.C, Sommerhalter, M, Baldwin, J, Saleh, L, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-06-09 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Variable coordination geometries at the diiron(II) active site of ribonucleotide reductase R2.
J.Am.Chem.Soc., 125, 2003
|
|
5TWK
 
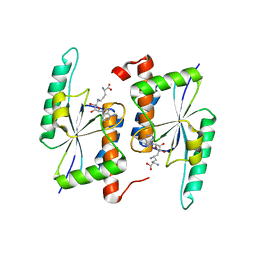 | | Crystal Structure of RlmH in Complex with Sinefungin | | Descriptor: | Ribosomal RNA large subunit methyltransferase H, SINEFUNGIN | | Authors: | Koh, C.S, Madireddy, R, Beane, T.J, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small methyltransferase RlmH assembles a composite active site to methylate a ribosomal pseudouridine.
Sci Rep, 7, 2017
|
|
6DNC
 
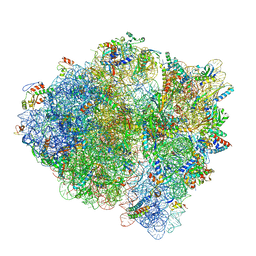 | | E.coli RF1 bound to E.coli 70S ribosome in response to UAU sense A-site codon | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Demo, G, Korostelev, A.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of premature translation termination on a sense codon.
J. Biol. Chem., 293, 2018
|
|
5ELG
 
 | |
5IN7
 
 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-07 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
1YJU
 
 | | Solution structure of the apo form of the sixth soluble domain of Menkes protein | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Rosato, A, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-15 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An atomic-level investigation of the disease-causing A629P mutant of the Menkes protein, ATP7A
J.Mol.Biol., 352, 2005
|
|
1YJR
 
 | | Solution structure of the apo form of the sixth soluble domain A69P mutant of Menkes protein | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Rosato, A, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-15 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An atomic-level investigation of the disease-causing A629P mutant of the Menkes protein, ATP7A
J.Mol.Biol., 352, 2005
|
|
3RFA
 
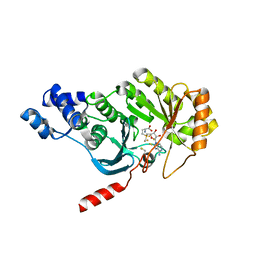 | | X-ray structure of RlmN from Escherichia coli in complex with S-adenosylmethionine | | Descriptor: | IRON/SULFUR CLUSTER, Ribosomal RNA large subunit methyltransferase N, S-ADENOSYLMETHIONINE | | Authors: | Boal, A.K, Grove, T.L, McLaughlin, M.I, Yennawar, N, Booker, S.J, Rosenzweig, A.C. | | Deposit date: | 2011-04-05 | | Release date: | 2011-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for methyl transfer by a radical SAM enzyme.
Science, 332, 2011
|
|
1Q19
 
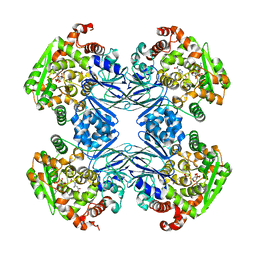 | | Carbapenam Synthetase | | Descriptor: | (2S,5S)-5-CARBOXYMETHYLPROLINE, CarA, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Miller, M.T, Gerratana, B, Stapon, A, Townsend, C.A, Rosenzweig, A.C. | | Deposit date: | 2003-07-18 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Carbapenam Synthetase (CarA)
J.Biol.Chem., 278, 2003
|
|
2WK0
 
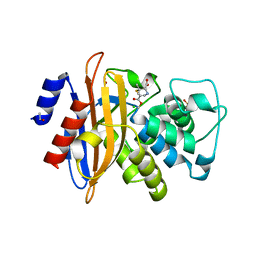 | | Crystal structure of the class A beta-lactamase BS3 inhibited by 6- beta-iodopenicillanate. | | Descriptor: | (3S)-2,2-dimethyl-3,4-dihydro-2H-1,4-thiazine-3,6-dicarboxylic acid, BETA-LACTAMASE, CHLORIDE ION, ... | | Authors: | Sauvage, E, Zervosen, A, Dive, G, Herman, R, Kerff, F, Amoroso, A, Fonze, E, Pratt, R.F, Luxen, A, Charlier, P. | | Deposit date: | 2009-06-03 | | Release date: | 2009-12-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis of the Inhibition of Class a Beta-Lactamases and Penicillin-Binding Proteins by 6-Beta-Iodopenicillanate.
J.Am.Chem.Soc., 131, 2009
|
|
3RFR
 
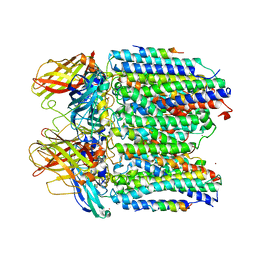 | |
3RF9
 
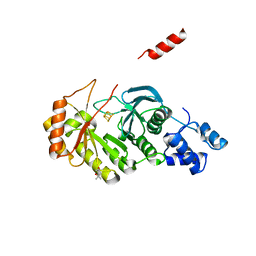 | | X-ray structure of RlmN from Escherichia coli | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, IRON/SULFUR CLUSTER, Ribosomal RNA large subunit methyltransferase N | | Authors: | Boal, A.K, Grove, T.L, McLaughlin, M.I, Yennawar, N, Booker, S.J, Rosenzweig, A.C. | | Deposit date: | 2011-04-05 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for methyl transfer by a radical SAM enzyme.
Science, 332, 2011
|
|
1NOF
 
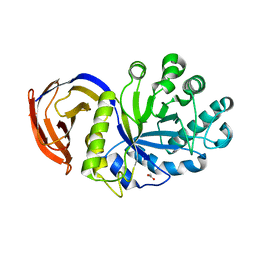 | | THE FIRST CRYSTALLOGRAPHIC STRUCTURE OF A XYLANASE FROM GLYCOSYL HYDROLASE FAMILY 5: IMPLICATIONS FOR CATALYSIS | | Descriptor: | ACETATE ION, xylanase | | Authors: | Larson, S.B, Day, J, McPherson, A, Barba De La Rosa, A.P, Keen, N.T. | | Deposit date: | 2003-01-16 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | First crystallographic structure of a xylanase from glycoside hydrolase family 5: implications for catalysis.
Biochemistry, 42, 2003
|
|
4PI0
 
 | |
2B7K
 
 | |
5WP9
 
 | | Structural Basis of Mitochondrial Receptor Binding and Constriction by Dynamin-Related Protein 1 | | Descriptor: | Dynamin-1-like protein, MAGNESIUM ION, Mitochondrial dynamics protein MID49, ... | | Authors: | Kalia, R, Wang, R.Y.R, Yusuf, A, Thomas, P.V, Agard, D.A, Shaw, J.M, Frost, A. | | Deposit date: | 2017-08-03 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Structural basis of mitochondrial receptor binding and constriction by DRP1.
Nature, 558, 2018
|
|
1URK
 
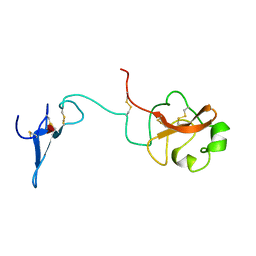 | | SOLUTION STRUCTURE OF THE AMINO TERMINAL FRAGMENT OF UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | PLASMINOGEN ACTIVATOR, alpha-L-fucopyranose | | Authors: | Hansen, A.P, Petros, A.M, Meadows, R.P, Nettesheim, D.G, Mazar, A.P, Olejniczak, E.T, Xu, R.X, Pederson, T.M, Henkin, J, Fesik, S.W. | | Deposit date: | 1994-01-10 | | Release date: | 1995-05-08 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the amino-terminal fragment of urokinase-type plasminogen activator.
Biochemistry, 33, 1994
|
|
2B4J
 
 | | Structural basis for the recognition between HIV-1 integrase and LEDGF/p75 | | Descriptor: | GLYCEROL, Integrase (IN), PC4 and SFRS1 interacting protein, ... | | Authors: | Cherepanov, P, Ambrosio, A.L, Rahman, S, Ellenberger, T, Engelman, A. | | Deposit date: | 2005-09-24 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
6AWB
 
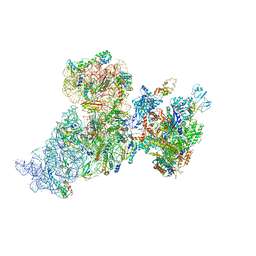 | | Structure of 30S ribosomal subunit and RNA polymerase complex in non-rotated state | | Descriptor: | 16S rRNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
3RJ6
 
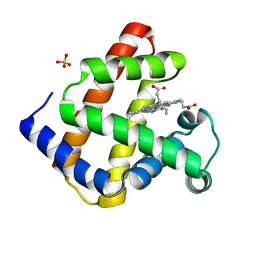 | |
6AWD
 
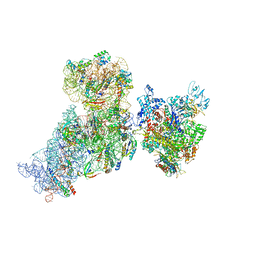 | | Structure of 30S (S1 depleted) ribosomal subunit and RNA polymerase complex | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
