5IN7
 
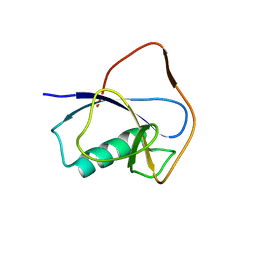 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-07 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
5JUU
 
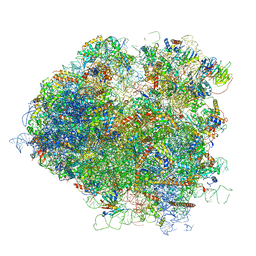 | | Saccharomyces cerevisiae 80S ribosome bound with elongation factor eEF2-GDP-sordarin and Taura Syndrome Virus IRES, Structure V (least rotated 40S subunit) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Abeyrathne, P, Koh, C.S, Grant, T, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-05 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome.
Elife, 5, 2016
|
|
5JUP
 
 | | Saccharomyces cerevisiae 80S ribosome bound with elongation factor eEF2-GDP-sordarin and Taura Syndrome Virus IRES, Structure II (mid-rotated 40S subunit) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Abeyrathne, P, Koh, C.S, Grant, T, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-05 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome.
Elife, 5, 2016
|
|
5TEG
 
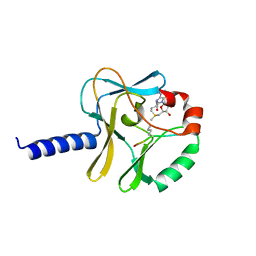 | |
5UYL
 
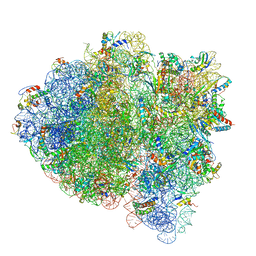 | | 70S ribosome bound with cognate ternary complex base-paired to A site codon (Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ensemble cryo-EM elucidates the mechanism of translation fidelity.
Nature, 546, 2017
|
|
5IOI
 
 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
5JUS
 
 | | Saccharomyces cerevisiae 80S ribosome bound with elongation factor eEF2-GDP-sordarin and Taura Syndrome Virus IRES, Structure III (mid-rotated 40S subunit) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Abeyrathne, P, Koh, C.S, Grant, T, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-05 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome.
Elife, 5, 2016
|
|
6AWC
 
 | | Structure of 30S ribosomal subunit and RNA polymerase complex in rotated state | | Descriptor: | 16S rRNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
6QT9
 
 | | Cryo-EM structure of SH1 full particle. | | Descriptor: | ORF 24, ORF 25, ORF 31, ... | | Authors: | De Colibus, L, Roine, E, Walter, T.S, Ilca, S.L, Wang, X, Wang, N, Roseman, A.M, Bamford, D, Huiskonen, J.T, Stuart, D.I. | | Deposit date: | 2019-02-22 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Assembly of complex viruses exemplified by a halophilic euryarchaeal virus.
Nat Commun, 10, 2019
|
|
5DOY
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with antibiotic Hygromycin A, mRNA and three tRNAs in the A, P and E sites at 2.6A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Starosta, A.L, Juette, M.F, Altman, R.B, Terry, D.S, Lu, W, Burnett, B.J, Dinos, G, Reynolds, K, Blanchard, S.C, Steitz, T.A, Wilson, D.N. | | Deposit date: | 2015-09-11 | | Release date: | 2015-12-30 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A.
Mol.Cell, 58, 2015
|
|
2B4J
 
 | | Structural basis for the recognition between HIV-1 integrase and LEDGF/p75 | | Descriptor: | GLYCEROL, Integrase (IN), PC4 and SFRS1 interacting protein, ... | | Authors: | Cherepanov, P, Ambrosio, A.L, Rahman, S, Ellenberger, T, Engelman, A. | | Deposit date: | 2005-09-24 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3ZM8
 
 | | Crystal structure of Podospora anserina GH26-CBM35 beta-(1,4)- mannanase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GH26 ENDO-BETA-1,4-MANNANASE, ... | | Authors: | Couturier, M, Roussel, A, Rosengren, A, Leone, P, Stalbrand, H, Berrin, J.G. | | Deposit date: | 2013-02-06 | | Release date: | 2013-04-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and Biochemical Analyses of Glycoside Hydrolase Families 5 and 26 Beta-(1,4)-Mannanases from Podospora Anserina Reveal Differences Upon Manno-Oligosaccharides Catalysis.
J.Biol.Chem., 288, 2013
|
|
5UYM
 
 | | 70S ribosome bound with cognate ternary complex base-paired to A site codon, closed 30S (Structure III) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ensemble cryo-EM elucidates the mechanism of translation fidelity
Nature, 546, 2017
|
|
5ELG
 
 | |
1H4M
 
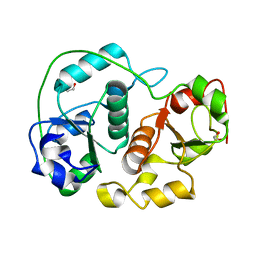 | | Sulfurtransferase from Azotobacter vinelandii in complex with phosphate | | Descriptor: | 1,2-ETHANEDIOL, PUTATIVE THIOSULFATE SULFURTRANSFERASE | | Authors: | Bordo, D, Forlani, F, Spallarossa, A, Colnaghi, R, Carpen, A, Pagani, S, Bolognesi, M. | | Deposit date: | 2001-05-11 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Persulfurated Cysteine Promotes Active Site Reactivity in Azotobacter Vinelandii Rhodanse
Biol.Chem., 382, 2001
|
|
4S2U
 
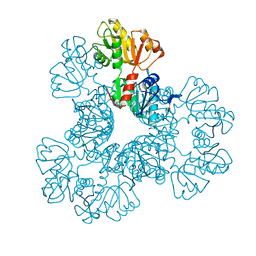 | | Crystal structure of the Phosphorybosylpyrophosphate synthetase from E. Coli | | Descriptor: | MAGNESIUM ION, Ribose-phosphate pyrophosphokinase | | Authors: | Timofeev, V.I, Abramchik, Y.A, Muravieva, T.I, Iaroslavtceva, A.K, Stepanenko, V.N, Zhukhlistova, N.E, Esipov, R.S, Kuranova, I.P. | | Deposit date: | 2015-01-23 | | Release date: | 2016-01-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of the Phosphorybosylpyrophosphate synthetase from E. Coli
To be Published
|
|
1H4K
 
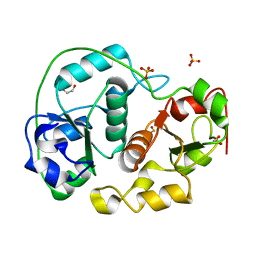 | | Sulfurtransferase from Azotobacter vinelandii in complex with hypophosphite | | Descriptor: | 1,2-ETHANEDIOL, HYPOPHOSPHITE, SULFATE ION, ... | | Authors: | Bordo, D, Forlani, F, Spallarossa, A, Colnaghi, R, Carpen, A, Pagani, S, Bolognesi, M. | | Deposit date: | 2001-05-11 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Persulfurated Cysteine Promotes Active Site Reactivity in Azotobacter Vinelandii Rhodanse
Biol.Chem., 382, 2001
|
|
6BU8
 
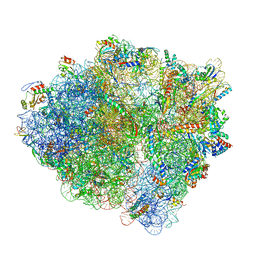 | | 70S ribosome with S1 domains 1 and 2 (Class 1) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Loveland, A.B, Korostelev, A.A. | | Deposit date: | 2017-12-08 | | Release date: | 2018-01-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural dynamics of protein S1 on the 70S ribosome visualized by ensemble cryo-EM.
Methods, 137, 2018
|
|
3TJB
 
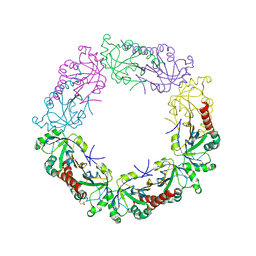 | | Crystal structure of wild-type human peroxiredoxin IV | | Descriptor: | Peroxiredoxin-4 | | Authors: | Cao, Z, Tavender, T.J, Roszak, A.W, Cogdell, R.J, Bulleid, N.J. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of Reduced and of Oxidized Peroxiredoxin IV Enzyme Reveals a Stable Oxidized Decamer and a Non-disulfide-bonded Intermediate in the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
3TJJ
 
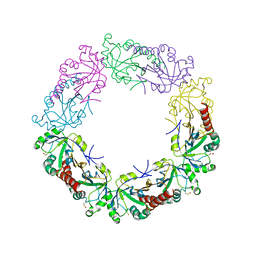 | | Crystal structure of human peroxiredoxin IV C245A mutant in sulfenylated form | | Descriptor: | Peroxiredoxin-4 | | Authors: | Cao, Z, Tavender, T.J, Roszak, A.W, Cogdell, R.J, Bulleid, N.J. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal Structure of Reduced and of Oxidized Peroxiredoxin IV Enzyme Reveals a Stable Oxidized Decamer and a Non-disulfide-bonded Intermediate in the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
6BOH
 
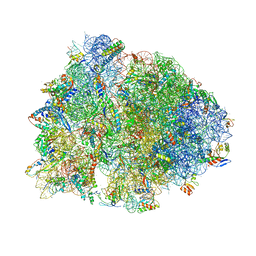 | |
3TJF
 
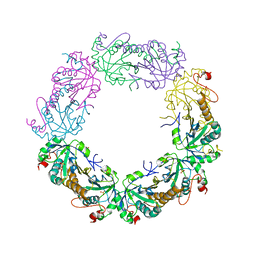 | | Crystal Structure of human peroxiredoxin IV C51A mutant in reduced form | | Descriptor: | Peroxiredoxin-4, SULFATE ION | | Authors: | Cao, Z, Tavender, T.J, Roszak, A.W, Cogdell, R.J, Bulleid, N.J. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal Structure of Reduced and of Oxidized Peroxiredoxin IV Enzyme Reveals a Stable Oxidized Decamer and a Non-disulfide-bonded Intermediate in the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
3ST1
 
 | | Crystal structure of Necrosis and Ethylene inducing Protein 2 from the causal agent of cocoa's Witches Broom disease | | Descriptor: | Necrosis-and ethylene-inducing protein, SODIUM ION, ZINC ION | | Authors: | Oliveira, J.F, Zaparoli, G, Barsottini, M.R.O, Ambrosio, A.L.B, Pereira, G.A.G, Dias, S.M.G. | | Deposit date: | 2011-07-08 | | Release date: | 2011-11-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Necrosis- and Ethylene-Inducing Protein 2 from the Causal Agent of Cacao's Witches' Broom Disease Reveals Key Elements for Its Activity.
Biochemistry, 50, 2011
|
|
1URK
 
 | | SOLUTION STRUCTURE OF THE AMINO TERMINAL FRAGMENT OF UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | PLASMINOGEN ACTIVATOR, alpha-L-fucopyranose | | Authors: | Hansen, A.P, Petros, A.M, Meadows, R.P, Nettesheim, D.G, Mazar, A.P, Olejniczak, E.T, Xu, R.X, Pederson, T.M, Henkin, J, Fesik, S.W. | | Deposit date: | 1994-01-10 | | Release date: | 1995-05-08 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the amino-terminal fragment of urokinase-type plasminogen activator.
Biochemistry, 33, 1994
|
|
2B7K
 
 | |
