2C7E
 
 | | REVISED ATOMIC STRUCTURE FITTING INTO A GROEL(D398A)-ATP7 CRYO-EM MAP (EMD 1047) | | Descriptor: | 60 KDA CHAPERONIN, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ranson, N.A, Farr, G.W, Roseman, A.M, Gowen, B, Fenton, W.A, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2005-11-22 | | Release date: | 2006-02-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (14.9 Å) | | Cite: | ATP-Bound States of Groel Captured by Cryo-Electron Microscopy
Cell(Cambridge,Mass.), 107, 2001
|
|
3ST1
 
 | | Crystal structure of Necrosis and Ethylene inducing Protein 2 from the causal agent of cocoa's Witches Broom disease | | Descriptor: | Necrosis-and ethylene-inducing protein, SODIUM ION, ZINC ION | | Authors: | Oliveira, J.F, Zaparoli, G, Barsottini, M.R.O, Ambrosio, A.L.B, Pereira, G.A.G, Dias, S.M.G. | | Deposit date: | 2011-07-08 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Necrosis- and Ethylene-Inducing Protein 2 from the Causal Agent of Cacao's Witches' Broom Disease Reveals Key Elements for Its Activity.
Biochemistry, 50, 2011
|
|
4NS2
 
 | |
3SUM
 
 | | Crystal structure of cerato-platanin 5 from M. perniciosa (MpCP5) | | Descriptor: | Cerato-platanin-like protein | | Authors: | Oliveira, J.F, Barsottini, M.R.O, Zaparoli, G, Machado, L.O, Dias, S.M.G, Pereira, G.A.G, Ambrosio, A.L.B. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization.
Mol. Plant Microbe Interact., 26, 2013
|
|
5UYK
 
 | | 70S ribosome bound with cognate ternary complex not base-paired to A site codon (Structure I) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ensemble cryo-EM elucidates the mechanism of translation fidelity
Nature, 546, 2017
|
|
5UYP
 
 | | 70S ribosome bound with near-cognate ternary complex base-paired to A site codon, open 30S (Structure II-nc) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ensemble cryo-EM elucidates the mechanism of translation fidelity
Nature, 546, 2017
|
|
3IFX
 
 | | Crystal structure of the Spin-labeled KcsA mutant V48R1 | | Descriptor: | POTASSIUM ION, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, TETRABUTYLAMMONIUM ION, ... | | Authors: | Cieslak, J.A, Focia, P.J, Gross, A. | | Deposit date: | 2009-07-26 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | EPR (3.56 Å), X-RAY DIFFRACTION | | Cite: | Electron Spin-Echo Envelope Modulation (ESEEM) Reveals Water and Phosphate Interactions with the KcsA Potassium Channel
Biochemistry, 49, 2010
|
|
1FE4
 
 | | CRYSTAL STRUCTURE OF MERCURY-HAH1 | | Descriptor: | COPPER TRANSPORT PROTEIN ATOX1, MERCURY (II) ION, SULFATE ION, ... | | Authors: | Wernimont, A.K, Huffman, D.L, Lamb, A.L, O'Halloran, T.V, Rosenzweig, A.C. | | Deposit date: | 2000-07-20 | | Release date: | 2001-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for copper transfer by the metallochaperone for the Menkes/Wilson disease proteins.
Nat.Struct.Biol., 7, 2000
|
|
1FEE
 
 | | CRYSTAL STRUCTURE OF COPPER-HAH1 | | Descriptor: | COPPER (I) ION, COPPER TRANSPORT PROTEIN ATOX1, SULFATE ION, ... | | Authors: | Wernimont, A.K, Huffman, D.L, Lamb, A.L, O'Halloran, T.V, Rosenzweig, A.C. | | Deposit date: | 2000-07-21 | | Release date: | 2001-01-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for copper transfer by the metallochaperone for the Menkes/Wilson disease proteins.
Nat.Struct.Biol., 7, 2000
|
|
3TG7
 
 | | Crystal structure of Adenovirus serotype 5 hexon at 1.6A resolution | | Descriptor: | Hexon protein | | Authors: | Zhu, Y, Roszak, A.W, Isaacs, N.W, McVey, J.H, Nicklin, S.A, Baker, A.H. | | Deposit date: | 2011-08-17 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | crystal structure of Adenovirus serotype 5 hexon at 1.6A resolution
To be Published
|
|
2WS2
 
 | |
1HKS
 
 | | SOLUTION STRUCTURE OF THE DNA-BINDING DOMAIN OF DROSOPHILA HEAT SHOCK TRANSCRIPTION FACTOR | | Descriptor: | HEAT-SHOCK TRANSCRIPTION FACTOR | | Authors: | Vuister, G.W, Kim, S.-J, Orosz, A, Marquardt, J.L, Wu, C, Bax, A. | | Deposit date: | 1994-07-18 | | Release date: | 1994-09-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of Drosophila heat shock transcription factor.
Nat.Struct.Biol., 1, 1994
|
|
1HKT
 
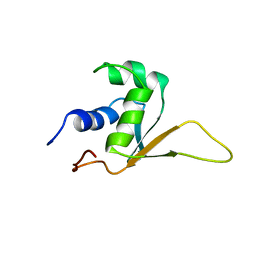 | | SOLUTION STRUCTURE OF THE DNA-BINDING DOMAIN OF DROSOPHILA HEAT SHOCK TRANSCRIPTION FACTOR | | Descriptor: | HEAT-SHOCK TRANSCRIPTION FACTOR | | Authors: | Vuister, G.W, Kim, S.-J, Orosz, A, Marquardt, J.L, Wu, C, Bax, A. | | Deposit date: | 1994-07-18 | | Release date: | 1994-09-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of Drosophila heat shock transcription factor.
Nat.Struct.Biol., 1, 1994
|
|
5D8B
 
 | |
3F8Z
 
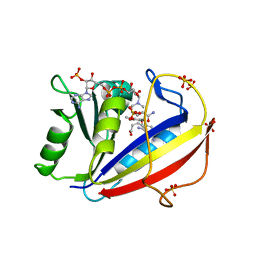 | | Human Dihydrofolate Reductase Structural Data with Active Site Mutant Enzyme Complexes | | Descriptor: | 2,4-DIAMINO-5-[2-METHOXY-5-(4-CARBOXYBUTYLOXY)BENZYL]PYRIMIDINE, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V, Pace, J, Makin, J, Piraino, J, Queener, S.F, Rosowsky, A. | | Deposit date: | 2008-11-13 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Correlations of Inhibitor Kinetics for Pneumocystis jirovecii and Human Dihydrofolate Reductase with Structural Data for Human Active Site Mutant Enzyme Complexes.
Biochemistry, 48, 2009
|
|
5DOX
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Hygromycin-A at 3.1A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Starosta, A.L, Juette, M.F, Altman, R.B, Terry, D.S, Lu, W, Burnett, B.J, Dinos, G, Reynolds, K, Blanchard, S.C, Steitz, T.A, Wilson, D.N. | | Deposit date: | 2015-09-11 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A.
Mol.Cell, 58, 2015
|
|
5U9F
 
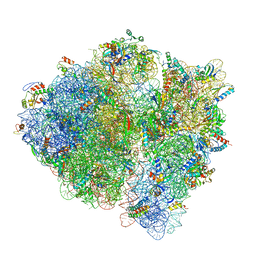 | | 3.2 A cryo-EM ArfA-RF2 ribosome rescue complex (Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Svidritskiy, E, Madireddy, R, Diaz-Avalos, R, Grant, T, Grigorieff, N, Sousa, D, Korostelev, A.A. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of ribosome rescue by ArfA and RF2.
Elife, 6, 2017
|
|
1KX7
 
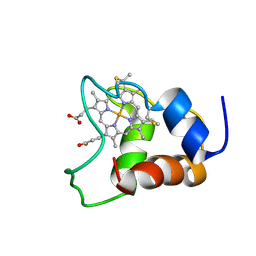 | | Family of 30 conformers of a mono-heme ferrocytochrome c from Shewanella putrefaciens solved by NMR | | Descriptor: | HEME C, mono-heme c-type cytochrome ScyA | | Authors: | Bartalesi, I, Bertini, I, Hajieva, P, Rosato, A, Vasos, P.R. | | Deposit date: | 2002-01-31 | | Release date: | 2002-02-13 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a monoheme ferrocytochrome c from Shewanella putrefaciens and structural analysis of sequence-similar proteins: functional implications.
Biochemistry, 41, 2002
|
|
4H49
 
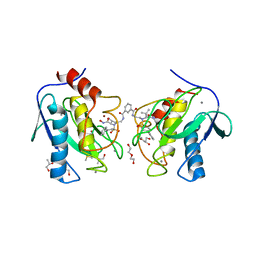 | | Crystal structure of the catalytic domain of MMP-12 in complex with a twin inhibitor. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Antoni, C, Stura, E.A, Vera, L, Nuti, E, Carafa, L, Cassar-Lajeunesse, E, Dive, V, Rossello, A. | | Deposit date: | 2012-09-17 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystallization of bi-functional ligand protein complexes.
J.Struct.Biol., 182, 2013
|
|
1JK0
 
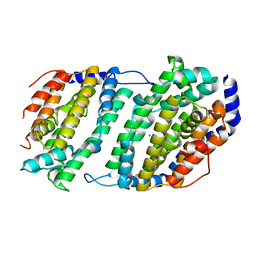 | | Ribonucleotide reductase Y2Y4 heterodimer | | Descriptor: | ZINC ION, ribonucleoside-diphosphate reductase small chain 1, ribonucleoside-diphosphate reductase small chain 2 | | Authors: | Voegtli, W.C, Perlstein, D.L, Ge, J, Stubbe, J, Rosenzweig, A.C. | | Deposit date: | 2001-07-10 | | Release date: | 2001-09-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the yeast ribonucleotide reductase Y2Y4 heterodimer.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
5UYM
 
 | | 70S ribosome bound with cognate ternary complex base-paired to A site codon, closed 30S (Structure III) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ensemble cryo-EM elucidates the mechanism of translation fidelity
Nature, 546, 2017
|
|
5UYQ
 
 | | 70S ribosome bound with near-cognate ternary complex base-paired to A site codon, closed 30S (Structure III-nc) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ensemble cryo-EM elucidates the mechanism of translation fidelity
Nature, 546, 2017
|
|
3IWX
 
 | |
1FZI
 
 | | METHANE MONOOXYGENASE HYDROXYLASE, FORM I PRESSURIZED WITH XENON GAS | | Descriptor: | FE (III) ION, METHANE MONOOXYGENASE COMPONENT A, ALPHA CHAIN, ... | | Authors: | Whittington, D.A, Rosenzweig, A.C, Frederick, C.A, Lippard, S.J. | | Deposit date: | 2000-10-03 | | Release date: | 2001-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Xenon and halogenated alkanes track putative substrate binding cavities in the soluble methane monooxygenase hydroxylase.
Biochemistry, 40, 2001
|
|
2XPC
 
 | | Second-generation sulfonamide inhibitors of MurD: Activity optimisation with conformationally rigid analogues of D-glutamic acid | | Descriptor: | (1R,3R,4S)-4-[({6-[(4-CYANO-2-FLUOROBENZYL)OXY]NAPHTHALEN-2-YL}SULFONYL)AMINO]CYCLOHEXANE-1,3-DICARBOXYLIC ACID, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Sosic, I, Barreteau, H, Simcic, M, Sink, R, Cesar, J, Golic-Grdadolnik, S, Contreras-Martel, C, Dessen, A, Amoroso, A, Joris, B, Blanot, D, Gobec, S. | | Deposit date: | 2010-08-26 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Second-Generation Sulfonamide Inhibitors of D- Glutamic Acid-Adding Enzyme: Activity Optimisation with Conformationally Rigid Analogues of D- Glutamic Acid.
Eur.J.Med.Chem, 46, 2011
|
|
