7ADP
 
 | | Cryo-EM structure of human ER membrane protein complex in GDN detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ER membrane protein complex subunit 10, ... | | Authors: | Braeuning, B, Prabu, J.R, Miller-Vedam, L.E, Weissman, J.S, Frost, A, Schulman, B.A. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
7A2D
 
 | | Structure-function analyses of dual-BON domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation to the cell division site | | Descriptor: | Uncharacterized protein YraP | | Authors: | Bryant, J.A, Morris, F.C, Knowles, T.J, Maderbocus, R, Heinz, E, Boelter, G, Alodaini, D, Colyer, A, Wotherspoon, P.J, Staunton, K.A, Jeeves, M, Browning, D.F, Sevastsyanovich, Y.R, Wells, T.J, Rossiter, A.E, Bavro, V.N, Sridhar, P, Ward, D.G, Chong, Z.S, Goodall, E.C.A, Icke, C, Teo, A, Chng, S.S, Roper, D.I, Lithgow, T, Cunningham, A.F, Banzhaf, M, Overduin, M, Henderson, I.R. | | Deposit date: | 2020-08-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of dual BON-domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation.
Elife, 9, 2020
|
|
6WNW
 
 | | Active 70S ribosome without free 5S rRNA and bound with A- and P- tRNA | | Descriptor: | 16S ribosomal RNA, 23s-5s joint ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Korostelev, A.A, Mankin, A.S, Huang, S, Aleksashin, N.A, Klepacki, D, Reier, K, Kefi, A, Szal, A, Remme, J, Jaeger, L, Vazquez-Laslop, N. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ribosome engineering reveals the importance of 5S rRNA autonomy for ribosome assembly.
Nat Commun, 11, 2020
|
|
6WNT
 
 | | 50S ribosomal subunit without free 5S rRNA and perturbed PTC | | Descriptor: | 23s-5s joint ribosomal RNA, 50S ribosomal protein L1, 50S ribosomal protein L10, ... | | Authors: | Loveland, A.B, Korostelev, A.A, Mankin, A.S, Huang, S, Aleksashin, N.A, Klepacki, D, Reier, K, Kefi, A, Szal, A, Remme, J, Jaeger, L, Vazquez-Laslop, N. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ribosome engineering reveals the importance of 5S rRNA autonomy for ribosome assembly.
Nat Commun, 11, 2020
|
|
2GGP
 
 | | Solution structure of the Atx1-Cu(I)-Ccc2a complex | | Descriptor: | COPPER (I) ION, Metal homeostasis factor ATX1, Probable copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Cantini, F, Felli, I.C, Gonnelli, L, Hadjiliadis, N, Pierattelli, R, Rosato, A, Voulgaris, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-24 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Atx1-Ccc2 complex is a metal-mediated protein-protein interaction.
Nat.Chem.Biol., 2, 2006
|
|
6WNV
 
 | | 70S ribosome without free 5S rRNA and with a perturbed PTC | | Descriptor: | 16S ribosomal RNA, 23s-5s joint ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Korostelev, A.A, Mankin, A.S, Huang, S, Aleksashin, N.A, Klepacki, D, Reier, K, Kefi, A, Szal, A, Remme, J, Jaeger, L, Vazquez-Laslop, N. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ribosome engineering reveals the importance of 5S rRNA autonomy for ribosome assembly.
Nat Commun, 11, 2020
|
|
2GA7
 
 | | Solution structure of the copper(I) form of the third metal-binding domain of ATP7A protein (menkes disease protein) | | Descriptor: | COPPER (I) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, DellaMalva, N, Rosato, A, Herrmann, T, Wuthrich, K, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-08 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and intermolecular interactions of the third metal-binding domain of ATP7A, the Menkes disease protein.
J.Biol.Chem., 281, 2006
|
|
1Q59
 
 | | Solution Structure of the BHRF1 Protein From Epstein-Barr Virus, a Homolog of Human Bcl-2 | | Descriptor: | Early antigen protein R | | Authors: | Huang, Q, Petros, A.M, Virgin, H.W, Fesik, S.W, Olejniczak, E.T. | | Deposit date: | 2003-08-06 | | Release date: | 2003-09-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the BHRF1 Protein From Epstein-Barr Virus, a Homolog of Human Bcl-2
J.Mol.Biol., 332, 2003
|
|
2FZI
 
 | | New Insights into DHFR Interactions: Analysis of Pneumocystis carinii and Mouse DHFR Complexes with NADPH and Two Highly Potent Trimethoprim Derivatives | | Descriptor: | 2,4-DIAMINO-5-[3',4'-DIMETHOXY-5'-(5-CARBOXYL-1-PENTYNYL)]BENZYL PYRIMIDINE, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Pace, J, Chisum, K, Rosowsky, A. | | Deposit date: | 2006-02-09 | | Release date: | 2006-12-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New insights into DHFR interactions: analysis of Pneumocystis carinii and mouse DHFR complexes with NADPH and two highly potent 5-(omega-carboxy(alkyloxy) trimethoprim derivatives reveals conformational correlations with activity and novel parallel ring stacking interactions.
Proteins, 65, 2006
|
|
2FZH
 
 | | New Insights into Dihydrofolate Reductase Interactions: Analysis of Pneumocystis carinii and Mouse DHFR Complexes with NADPH and Two Highly Potent Trimethoprim Derivatives | | Descriptor: | 2,4-DIAMINO-5-[2-METHOXY-5-(4-CARBOXYBUTYLOXY)BENZYL]PYRIMIDINE, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Pace, J, Chisum, K, Rosowsky, A. | | Deposit date: | 2006-02-09 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New insights into DHFR interactions: analysis of Pneumocystis carinii and mouse DHFR complexes with NADPH and two highly potent 5-(omega-carboxy(alkyloxy) trimethoprim derivatives reveals conformational correlations with activity and novel parallel ring stacking interactions.
Proteins, 65, 2006
|
|
2FZJ
 
 | | New Insights into DHFR Interactions: Analysis of Pneumocystis carinii and Mouse DHFR Complexes with NADPH and Two Highly Potent Trimethoprim Derivatives | | Descriptor: | 2,4-DIAMINO-5-[3',4'-DIMETHOXY-5'-(5-CARBOXYL-1-PENTYNYL)]BENZYL PYRIMIDINE, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Pace, J, Chisum, K, Rosowsky, A. | | Deposit date: | 2006-02-09 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New insights into DHFR interactions: analysis of Pneumocystis carinii and mouse DHFR complexes with NADPH and two highly potent 5-(omega-carboxy(alkyloxy) trimethoprim derivatives reveals conformational correlations with activity and novel parallel ring stacking interactions.
Proteins, 65, 2006
|
|
6HLK
 
 | | Hijacking the Hijackers: Escherichia coli Pathogenicity Islands Redirect Helper Phage Packaging for Their Own Benefit. | | Descriptor: | Redirecting phage packaging protein C (RppC) | | Authors: | Penades, J.R, Bacarizo, J, Marina, A, Alqasmi, M, Fillol-Salom, A, Roszak, A.W, Ciges-Tomas, J.R. | | Deposit date: | 2018-09-11 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Hijacking the Hijackers: Escherichia coli Pathogenicity Islands Redirect Helper Phage Packaging for Their Own Benefit.
Mol.Cell, 75, 2019
|
|
6HSR
 
 | |
2P7C
 
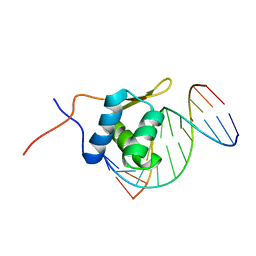 | | Solution structure of the bacillus licheniformis BlaI monomeric form in complex with the blaP half-operator. | | Descriptor: | Penicillinase repressor, Strand 1 of Twelve base-pair DNA, Strand 2 of Twelve base-pair DNA | | Authors: | Boudet, J, Duval, V, Van Melckebeke, H, Blackledge, M, Amoroso, A, Joris, B, Simorre, J.-P. | | Deposit date: | 2007-03-20 | | Release date: | 2007-06-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformational and thermodynamic changes of the repressor/DNA operator complex upon monomerization shed new light on regulation mechanisms of bacterial resistance against beta-lactam antibiotics.
Nucleic Acids Res., 35, 2007
|
|
3RJN
 
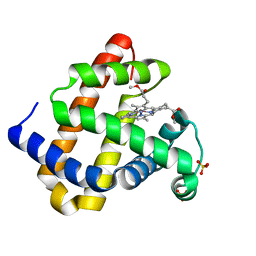 | |
1YSW
 
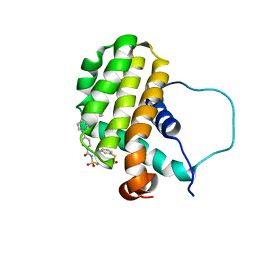 | | Solution structure of the anti-apoptotic protein Bcl-2 complexed with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-2 | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
1YSG
 
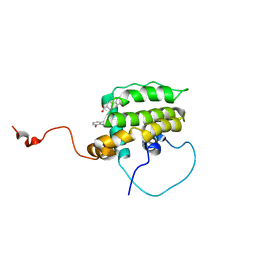 | | Solution Structure of the Anti-apoptotic Protein Bcl-xL in Complex with "SAR by NMR" Ligands | | Descriptor: | 4'-FLUORO-1,1'-BIPHENYL-4-CARBOXYLIC ACID, 5,6,7,8-TETRAHYDRONAPHTHALEN-1-OL, Apoptosis regulator Bcl-X | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-08 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
1YSI
 
 | | Solution structure of the anti-apoptotic protein Bcl-xL in complex with an acyl-sulfonamide-based ligand | | Descriptor: | Apoptosis regulator Bcl-X, N-[(4'-FLUORO-1,1'-BIPHENYL-4-YL)CARBONYL]-3-NITRO-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-08 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
1YSX
 
 | | Solution structure of domain 3 from human serum albumin complexed to an anti-apoptotic ligand directed against Bcl-xL and Bcl-2 | | Descriptor: | 4-({2-[(2,4-DIMETHYLPHENYL)SULFANYL]ETHYL}AMINO)-N-[(4'-FLUORO-1,1'-BIPHENYL-4-YL)CARBONYL]-3-NITROBENZENESULFONAMIDE, Serum albumin | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
2K1R
 
 | | The solution NMR structure of the complex between MNK1 and HAH1 mediated by Cu(I) | | Descriptor: | COPPER (II) ION, Copper transport protein ATOX1, Copper-transporting ATPase 1 | | Authors: | Bertini, I, Banci, L.C, Felli, I.C, Pavelkova, A, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2008-03-14 | | Release date: | 2009-03-31 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the copper(I)-mediated complex between the first soluble domain of the Menkes protein and the metallochaperone HAH1.
To be Published
|
|
2H2M
 
 | |
2PS3
 
 | | Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli | | Descriptor: | High-affinity zinc uptake system protein znuA | | Authors: | Yatsunyk, L.A, Kim, L.R, Vorontsov, I.I, Rosenzweig, A.C. | | Deposit date: | 2007-05-04 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli.
J.Biol.Inorg.Chem., 13, 2008
|
|
2PRS
 
 | | Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli | | Descriptor: | High-affinity zinc uptake system protein znuA, ISOPROPYL ALCOHOL, ZINC ION | | Authors: | Yatsunyk, L.A, Kim, L.R, Vorontsov, I.I, Rosenzweig, A.C. | | Deposit date: | 2007-05-04 | | Release date: | 2007-06-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli.
J.Biol.Inorg.Chem., 13, 2008
|
|
6P1F
 
 | | apo PmoF2 PCuAC domain | | Descriptor: | Copper chaperone PCu(A)C | | Authors: | Fisher, O.S, Sendzik, M.R, Rosenzweig, A.C. | | Deposit date: | 2019-05-19 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.654 Å) | | Cite: | PCuAC domains from methane-oxidizing bacteria use a histidine brace to bind copper.
J.Biol.Chem., 294, 2019
|
|
1SMS
 
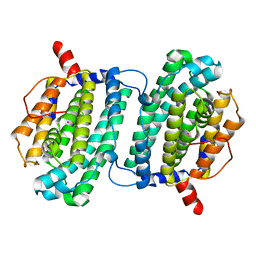 | | Structure of the Ribonucleotide Reductase Rnr4 Homodimer from Saccharomyces cerevisiae | | Descriptor: | MERCURY (II) ION, Ribonucleoside-diphosphate reductase small chain 2 | | Authors: | Sommerhalter, M, Voegtli, W.C, Perlstein, D.L, Ge, J, Stubbe, J, Rosenzweig, A.C. | | Deposit date: | 2004-03-09 | | Release date: | 2004-08-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the yeast ribonucleotide reductase Rnr2 and Rnr4 homodimers.
Biochemistry, 43, 2004
|
|
