7RYT
 
 | |
6H9U
 
 | | Crystal structure of the BiP NBD and MANF SAP complex | | Descriptor: | D-MALATE, Endoplasmic reticulum chaperone BiP, Mesencephalic astrocyte-derived neurotrophic factor, ... | | Authors: | Yan, Y, Ron, D. | | Deposit date: | 2018-08-06 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | MANF antagonizes nucleotide exchange by the endoplasmic reticulum chaperone BiP.
Nat Commun, 10, 2019
|
|
6HAB
 
 | | Crystal structure of BiP V461F (apo) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Endoplasmic reticulum chaperone BiP | | Authors: | Yan, Y, Ron, D. | | Deposit date: | 2018-08-07 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | MANF antagonizes nucleotide exchange by the endoplasmic reticulum chaperone BiP.
Nat Commun, 10, 2019
|
|
6HA7
 
 | | Crystal structure of the BiP NBD and MANF complex | | Descriptor: | 1,2-ETHANEDIOL, Endoplasmic reticulum chaperone BiP, Mesencephalic astrocyte-derived neurotrophic factor | | Authors: | Yan, Y, Ron, D. | | Deposit date: | 2018-08-07 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | MANF antagonizes nucleotide exchange by the endoplasmic reticulum chaperone BiP.
Nat Commun, 10, 2019
|
|
6I7J
 
 | | Crystal structure of monomeric FICD mutant L258D | | Descriptor: | Adenosine monophosphate-protein transferase FICD, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
6I7K
 
 | | Crystal structure of monomeric FICD mutant L258D complexed with MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenosine monophosphate-protein transferase FICD, ETHANOL, ... | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
6I7I
 
 | | Crystal structure of dimeric FICD mutant K256A complexed with MgATP | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ADENOSINE-5'-TRIPHOSPHATE, Adenosine monophosphate-protein transferase FICD, ... | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
6I7H
 
 | | Crystal structure of dimeric FICD mutant K256S | | Descriptor: | Adenosine monophosphate-protein transferase FICD, MAGNESIUM ION | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
6EOC
 
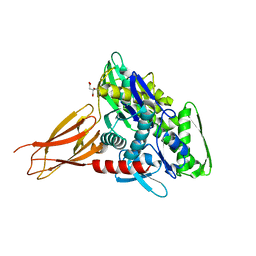 | | Crystal structure of AMPylated GRP78 in apo form (Crystal form 2) | | Descriptor: | 78 kDa glucose-regulated protein, CITRATE ANION, SULFATE ION | | Authors: | Yan, Y, Preissler, S, Ron, D, Read, R.J. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional inactivation.
Elife, 6, 2017
|
|
6EOF
 
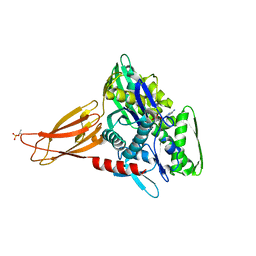 | | Crystal structure of AMPylated GRP78 in ADP state | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Yan, Y, Preissler, S, Read, R.J, Ron, D. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional inactivation.
Elife, 6, 2017
|
|
6EZO
 
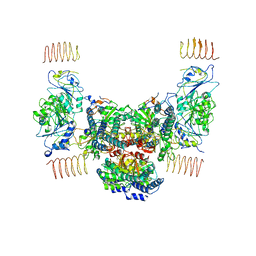 | | Eukaryotic initiation factor EIF2B in complex with ISRIB | | Descriptor: | 2-(4-chloranylphenoxy)-~{N}-[4-[2-(4-chloranylphenoxy)ethanoylamino]cyclohexyl]ethanamide, Human eukaryotic initiation factor EIF2B epsilon subunits, Translation initiation factor eIF-2B subunit alpha, ... | | Authors: | Faille, A, Weis, F, Zyryanova, A, Warren, A.J, Ron, D. | | Deposit date: | 2017-11-16 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Binding of ISRIB reveals a regulatory site in the nucleotide exchange factor eIF2B.
Science, 359, 2018
|
|
6EOE
 
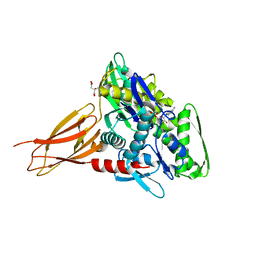 | | Crystal structure of AMPylated GRP78 with nucleotide | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE-5'-DIPHOSPHATE, CITRATE ANION, ... | | Authors: | Yan, Y, Preissler, S, Ron, D, Read, R.J. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional inactivation.
Elife, 6, 2017
|
|
5O4P
 
 | | Crystal structure of AMPylated GRP78 | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE MONOPHOSPHATE, SULFATE ION | | Authors: | Yan, Y, Chen, R, Ron, D, Read, R. | | Deposit date: | 2017-05-30 | | Release date: | 2017-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional inactivation.
Elife, 6, 2017
|
|
6EOB
 
 | | Crystal structure of AMPylated GRP78 in apo form (Crystal form 1) | | Descriptor: | 78 kDa glucose-regulated protein, PHOSPHATE ION | | Authors: | Yan, Y, Preissler, S, Ron, D, Read, R.J. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional inactivation.
Elife, 6, 2017
|
|
1NUN
 
 | | Crystal Structure Analysis of the FGF10-FGFR2b Complex | | Descriptor: | Fibroblast growth factor-10, POLYETHYLENE GLYCOL (N=34), SULFATE ION, ... | | Authors: | Yeh, B.K, Igarashi, M, Eliseenkova, A.V, Plotnikov, A.N, Sher, I, Ron, D, Aaronson, S.A, Mohammadi, M. | | Deposit date: | 2003-01-31 | | Release date: | 2003-02-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis by which alternative splicing confers
specificity in fibroblast growth factor receptors.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
6SHC
 
 | | Crystal structure of human IRE1 luminal domain Q105C | | Descriptor: | Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Yan, Y, Ron, D. | | Deposit date: | 2019-08-06 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Unstructured regions in IRE1 alpha specify BiP-mediated destabilisation of the luminal domain dimer and repression of the UPR.
Elife, 8, 2019
|
|
6ZYI
 
 | |
6ZYJ
 
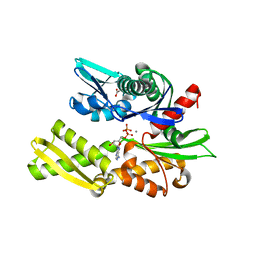 | | Crystal structure of Hsc70 ATPase domain in complex with ADP and calcium | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Yan, Y, Preissler, S, Ron, D. | | Deposit date: | 2020-08-01 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Calcium depletion challenges endoplasmic reticulum proteostasis by destabilising BiP-substrate complexes.
Elife, 9, 2020
|
|
7A4U
 
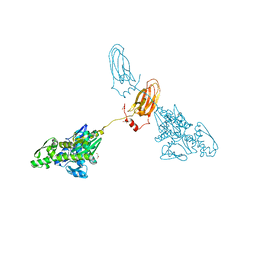 | |
6ZYH
 
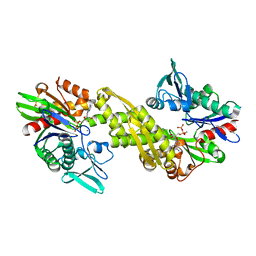 | |
7A4V
 
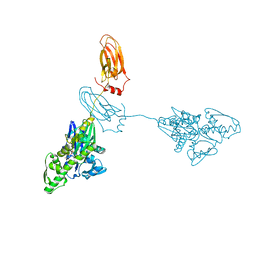 | |
5KWJ
 
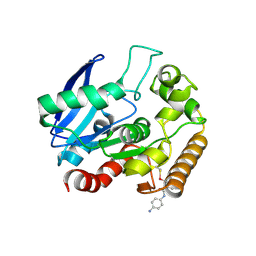 | | M.tb Ag85C modified at C209 by amino-ebselen | | Descriptor: | Diacylglycerol acyltransferase/mycolyltransferase Ag85C, ~{N}-(4-aminophenyl)-2-selanyl-benzamide | | Authors: | Goins, C.M, Ronning, D.R. | | Deposit date: | 2016-07-18 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Exploring Covalent Allosteric Inhibition of Antigen 85C from Mycobacterium tuberculosis by Ebselen Derivatives.
ACS Infect Dis, 3, 2017
|
|
7AEG
 
 | | SARS-CoV-2 main protease in a covalent complex with SDZ 224015 derivative, compound 5 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(benzyloxy)carbonyl]-L-valyl-N-[(1S)-1-(carboxymethyl)-3-fluoro-2-oxopropyl]-L-alaninamide | | Authors: | Owen, C.D, Redhead, M.A, Lukacik, P, Strain-Damerell, C, Fearon, D, Brewitz, L, Collette, A, Robinson, C, Collins, P, Radoux, C, Navratilova, I, Douangamath, A, von Delft, F, Malla, T.R, Nugen, T, Hull, H, Tumber, A, Schofield, C.J, Hallet, D, Stuart, D.I, Hopkins, A.L, Walsh, M.A. | | Deposit date: | 2020-09-17 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bispecific repurposed medicines targeting the viral and immunological arms of COVID-19.
Sci Rep, 11, 2021
|
|
7AEH
 
 | | SARS-CoV-2 main protease in a covalent complex with a pyridine derivative of ABT-957, compound 1 | | Descriptor: | (2~{R})-5-oxidanylidene-~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(pyridin-2-ylmethylamino)butan-2-yl]-1-(phenylmethyl)pyrrolidine-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Owen, C.D, Redhead, M.A, Lukacik, P, Strain-Damerell, C, Fearon, D, Brewitz, L, Collette, A, Robinson, C, Collins, P, Radoux, C, Navratilova, I, Douangamath, A, von Delft, F, Malla, T.R, Nugen, T, Hull, H, Tumber, A, Schofield, C.J, Hallet, D, Stuart, D.I, Hopkins, A.L, Walsh, M.A. | | Deposit date: | 2020-09-17 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Bispecific repurposed medicines targeting the viral and immunological arms of COVID-19.
Sci Rep, 11, 2021
|
|
4V41
 
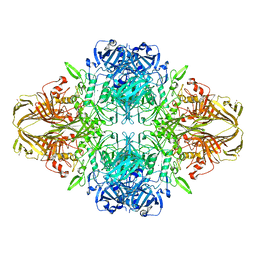 | | E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 2000-06-07 | | Release date: | 2014-07-09 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
