4H6Z
 
 | | Tubulin acetyltransferase | | Descriptor: | ACETYL COENZYME *A, Alpha-tubulin N-acetyltransferase, PHOSPHATE ION | | Authors: | Kizub, L, Szyk, A, Piszczek, G, Roll-Mecak, A. | | Deposit date: | 2012-09-19 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal structures of tubulin acetyltransferase reveal a conserved catalytic core and the plasticity of the essential N terminus.
J.Biol.Chem., 287, 2012
|
|
6P07
 
 | | Spastin hexamer in complex with substrate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sandate, C.R, Szyk, A, Zehr, E, Roll-Mecak, A, Lander, G.C. | | Deposit date: | 2019-05-16 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | An allosteric network in spastin couples multiple activities required for microtubule severing.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6QVJ
 
 | | HsCKK (human CAMSAP1) decorated 14pf taxol-GDP microtubule | | Descriptor: | Calmodulin-regulated spectrin-associated protein 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-03-02 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
6QUY
 
 | | NgCKK (N.Gruberi CKK) decorated 13pf taxol-GDP microtubule | | Descriptor: | CKK domain protein, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-02-28 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
6QVE
 
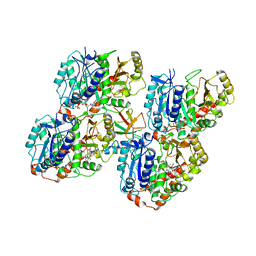 | | NgCKK (Naegleria Gruberi CKK) decorated 14pf taxol-GDP microtubule | | Descriptor: | Beta1-tubulin, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-03-01 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
1ZYC
 
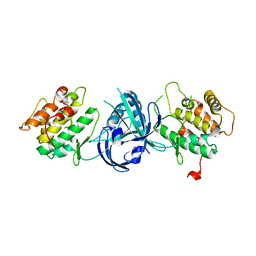 | | Crystal Structure of eIF2alpha Protein Kinase GCN2: Wild-Type in Apo Form. | | Descriptor: | Serine/threonine-protein kinase GCN2 | | Authors: | Padyana, A.K, Qiu, H, Roll-Mecak, A, Hinnebusch, A.G, Burley, S.K. | | Deposit date: | 2005-06-09 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Autoinhibition and Mutational Activation of Eukaryotic Initiation Factor 2{alpha} Protein Kinase GCN2
J.Biol.Chem., 280, 2005
|
|
8U3Z
 
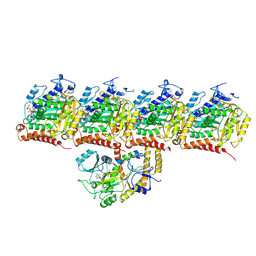 | | Model of TTLL6 bound to microtubule from composite map | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Mahalingan, K.K, Grotjahn, D, Li, Y, Lander, G.C, Zehr, E.A, Roll-Mecak, A. | | Deposit date: | 2023-09-08 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for alpha-tubulin-specific and modification state-dependent glutamylation.
Nat.Chem.Biol., 2024
|
|
8V4K
 
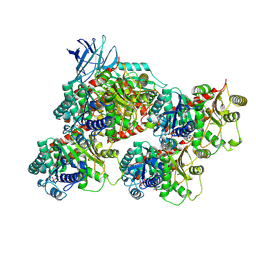 | | CCP5 in complex with microtubules class1 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-29 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3O
 
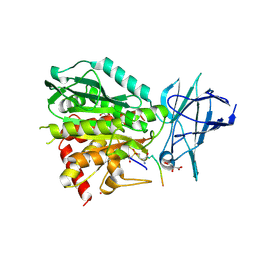 | | CCP5 in complex with Glu-P-peptide 1 transition state analog | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, D-MALATE, POTASSIUM ION, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V4M
 
 | | CCP5 in complex with microtubules class3 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-29 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3S
 
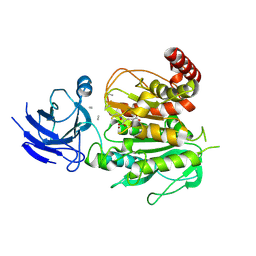 | | Structure of CCP5 class3 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, ZINC ION, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3M
 
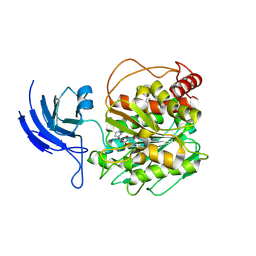 | | CCP5 apo structure | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, D-MALATE, IMIDAZOLE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3R
 
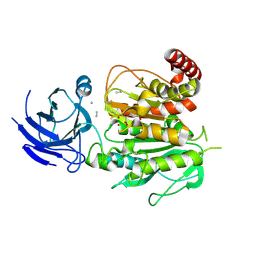 | | Structure of CCP5 class2 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, ZINC ION, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3P
 
 | | CCP5 in complex with Glu-P-peptide 2 transition state analog | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, Tubulin beta-2A chain, ZINC ION | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V4L
 
 | | CCP5 in complex with microtubules class2 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-29 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3Q
 
 | | Structure of CCP5 class1 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, ZINC ION, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
1ZY4
 
 | | Crystal Structure of eIF2alpha Protein Kinase GCN2: R794G Hyperactivating Mutant in Apo Form. | | Descriptor: | GLYCEROL, Serine/threonine-protein kinase GCN2 | | Authors: | Padyana, A.K, Qiu, H, Roll-Mecak, A, Hinnebusch, A.G, Burley, S.K. | | Deposit date: | 2005-06-09 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for Autoinhibition and Mutational Activation of Eukaryotic Initiation Factor 2{alpha} Protein Kinase GCN2
J.Biol.Chem., 280, 2005
|
|
1ZY5
 
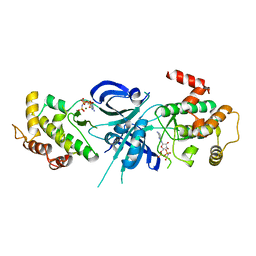 | | Crystal Structure of eIF2alpha Protein Kinase GCN2: R794G Hyperactivating Mutant Complexed with AMPPNP. | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase GCN2 | | Authors: | Padyana, A.K, Qiu, H, Roll-Mecak, A, Hinnebusch, A.G, Burley, S.K. | | Deposit date: | 2005-06-09 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Autoinhibition and Mutational Activation of Eukaryotic Initiation Factor 2{alpha} Protein Kinase GCN2
J.Biol.Chem., 280, 2005
|
|
1ZXE
 
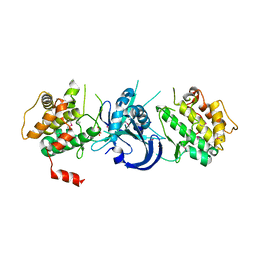 | | Crystal Structure of eIF2alpha Protein Kinase GCN2: D835N Inactivating Mutant in Apo Form | | Descriptor: | GLYCEROL, Serine/threonine-protein kinase | | Authors: | Padyana, A.K, Qiu, H, Roll-Mecak, A, Hinnebusch, A.G, Burley, S.K. | | Deposit date: | 2005-06-07 | | Release date: | 2005-06-21 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Autoinhibition and Mutational Activation of Eukaryotic Initiation Factor 2{alpha} Protein Kinase GCN2
J.Biol.Chem., 280, 2005
|
|
6UGF
 
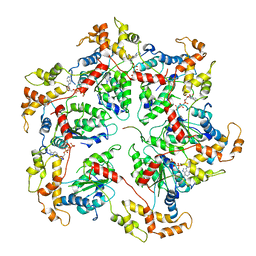 | |
8V3N
 
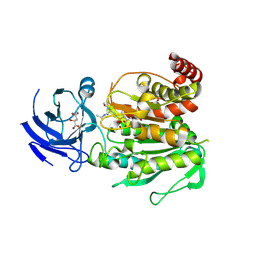 | | CCP5 in complex with Glu-P-Glu transition state analog | | Descriptor: | (2S)-2-{[(S)-[(3S)-3-acetamido-4-(ethylamino)-4-oxobutyl](hydroxy)phosphoryl]methyl}pentanedioic acid, Cytosolic carboxypeptidase-like protein 5, D-MALATE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
6VZQ
 
 | | Engineered TTLL6 mutant bound to alpha-elongation analog | | Descriptor: | (2~{S})-2-[[[(1~{R})-1-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, (2~{S})-2-[[[(1~{S})-1-acetamidoethyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, E.K, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VZS
 
 | | Engineered TTLL6 mutant bound to gamma-elongation analog | | Descriptor: | (2~{S})-2-[[[(3~{R})-3-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VZW
 
 | | TTLL6 bound to the initiation analog | | Descriptor: | (2~{S})-2-[[[(3~{R})-3-acetamido-4-(ethylamino)-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.L, Tanner, T.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VZU
 
 | | TTLL6 bound to alpha-elongation analog | | Descriptor: | (2~{S})-2-[[[(1~{R})-1-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-oxidanyl-phosphoryl]methyl]pentanedioic acid, (2~{S})-2-[[[(1~{R})-1-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.B, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
