4WRY
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-fluorouracil(B), Form I | | Descriptor: | 5-FLUOROURACIL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WPK
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase, Form I | | Descriptor: | CITRIC ACID, SODIUM ION, Uracil-DNA glycosylase | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-20 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1CR7
 
 | | PEANUT LECTIN-LACTOSE COMPLEX MONOCLINIC FORM | | Descriptor: | CALCIUM ION, LECTIN, MANGANESE (II) ION, ... | | Authors: | Ravishankar, R, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 1999-08-14 | | Release date: | 2001-04-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the peanut lectin-lactose complex at acidic pH: retention of unusual quaternary structure, empty and carbohydrate bound combining sites, molecular mimicry and crystal packing directed by interactions at the combining site.
Proteins, 43, 2001
|
|
1CQ9
 
 | | PEANUT LECTIN-TRICLINIC FORM | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PROTEIN (PEANUT LECTIN) | | Authors: | Ravishankar, R, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 1999-08-06 | | Release date: | 2002-05-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of the peanut lectin-lactose complex at acidic pH: retention of unusual
quaternary structure, empty and carbohydrate bound combining sites, molecular mimicry
and crystal packing directed by interactions at the combining site.
Proteins, 43, 2001
|
|
6XV0
 
 | | lauric acid functionalized hexamolybdoaluminate bound to human serum albumin | | Descriptor: | MYRISTIC ACID, Serum albumin, lauric acid functionalized hexamolybdoaluminate | | Authors: | Bijelic, A, Dobrov, A, Roller, A, Rompel, A. | | Deposit date: | 2020-01-21 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Binding of a Fatty Acid-Functionalized Anderson-Type Polyoxometalate to Human Serum Albumin.
Inorg.Chem., 59, 2020
|
|
1F9K
 
 | | WINGED BEAN ACIDIC LECTIN COMPLEXED WITH METHYL-ALPHA-D-GALACTOSE | | Descriptor: | ACIDIC LECTIN, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Manoj, N, Srinivas, V.R, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2000-07-11 | | Release date: | 2001-07-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Carbohydrate specificity and salt-bridge mediated conformational change in acidic winged bean agglutinin.
J.Mol.Biol., 302, 2000
|
|
1FAY
 
 | | WINGED BEAN ACIDIC LECTIN COMPLEXED WITH METHYL-ALPHA-D-GALACTOSE (MONOCLINIC FORM) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACIDIC LECTIN, CALCIUM ION, ... | | Authors: | Manoj, N, Srinivas, V.R, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2000-07-14 | | Release date: | 2001-07-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Carbohydrate specificity and salt-bridge mediated conformational change in acidic winged bean agglutinin.
J.Mol.Biol., 302, 2000
|
|
4QHG
 
 | | Crystal structure of Methanocaldococcus jannaschii dimeric selecase | | Descriptor: | GLYCEROL, Uncharacterized protein MJ1213, ZINC ION | | Authors: | Lopez-pelegrin, M, Cerda-costa, N, Cintas-pedrola, A, Herranz-trillo, F, Bernado, P, Peinado, J.R, Arolas, J.L, Gomis-ruth, F.X. | | Deposit date: | 2014-05-28 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple stable conformations account for reversible concentration-dependent oligomerization and autoinhibition of a metamorphic metallopeptidase
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4QHH
 
 | | Crystal structure of Methanocaldococcus jannaschii tetrameric selecase | | Descriptor: | GLYCEROL, SODIUM ION, Uncharacterized protein MJ1213, ... | | Authors: | Lopez-pelegrin, M, Cerda-costa, N, Cintas-pedrola, A, Herranz-trillo, F, Bernado, P, Peinado, J.R, Arolas, J.L, Gomis-ruth, F.X. | | Deposit date: | 2014-05-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Multiple stable conformations account for reversible concentration-dependent oligomerization and autoinhibition of a metamorphic metallopeptidase
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6Y2K
 
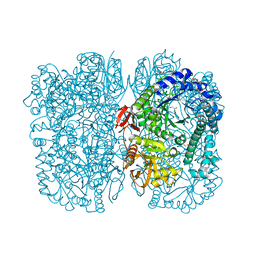 | | Crystal structure of beta-galactosidase from the psychrophilic Marinomonas ef1 | | Descriptor: | CHLORIDE ION, GLYCEROL, beta-galactosidase | | Authors: | Mangiagalli, M, Lapi, M, Maione, S, Orlando, M, Brocca, S, Pesce, A, Barbiroli, A, Pucciarelli, S, Camilloni, C, Lotti, M. | | Deposit date: | 2020-02-16 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The co-existence of cold activity and thermal stability in an Antarctic GH42 beta-galactosidase relies on its hexameric quaternary arrangement.
Febs J., 288, 2021
|
|
5L8S
 
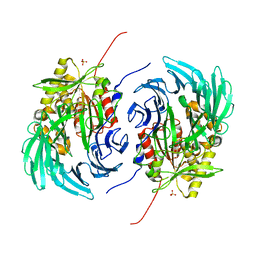 | | The crystal structure of a cold-adapted acylaminoacyl peptidase reveals a novel quaternary architecture based on the arm-exchange mechanism | | Descriptor: | Amino acyl peptidase, SULFATE ION | | Authors: | Brocca, S, Ferrari, C, Barbiroli, A, Pesce, A, Lotti, M, Nardini, M. | | Deposit date: | 2016-06-08 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A bacterial acyl aminoacyl peptidase couples flexibility and stability as a result of cold adaptation.
FEBS J., 283, 2016
|
|
5UZB
 
 | | Cryo-EM structure of the MAL TIR domain filament | | Descriptor: | Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Ve, T, Vajjhala, P.R, Hedger, A, Croll, T, DiMaio, F, Horsefield, S, Yu, X, Lavrencic, P, Hassan, Z, Morgan, G.P, Mansell, A, Mobli, M, O'Carrol, A, Chauvin, B, Gambin, Y, Sierecki, E, Landsberg, M.J, Stacey, K.J, Egelman, E.H, Kobe, B. | | Deposit date: | 2017-02-25 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis of TIR-domain-assembly formation in MAL- and MyD88-dependent TLR4 signaling.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7Z0H
 
 | | Structure of yeast RNA Polymerase III-Ty1 integrase complex at 2.6 A (focus subunit AC40). | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Nguyen, P.Q, Huecas, S, Plaza-Pegueroles, A, Fernandez-Tornero, C. | | Deposit date: | 2022-02-22 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of Ty1 integrase tethering to RNA polymerase III for targeted retrotransposon integration.
Nat Commun, 14, 2023
|
|
7Z31
 
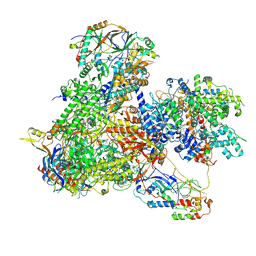 | | Structure of yeast RNA Polymerase III-Ty1 integrase complex at 2.7 A (focus subunit C11, no C11 C-terminal Zn-ribbon in the funnel pore). | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Nguyen, P.Q, Huecas, S, Plaza-Pegueroles, A, Fernandez-Tornero, C. | | Deposit date: | 2022-03-01 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis of Ty1 integrase tethering to RNA polymerase III for targeted retrotransposon integration.
Nat Commun, 14, 2023
|
|
4JIX
 
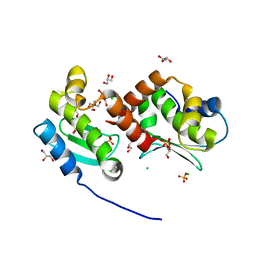 | | Crystal structure of the metallopeptidase zymogen of Methanocaldococcus jannaschii jannalysin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Lopez-Pelegrin, M, Cerda-Costa, N, Martinez-Jimenez, F, Cintas-Pedrola, A, Canals, A, Peinado, J.R, Marti-Renom, M.A, Lopez-Otin, C, Arolas, J.L, Gomis-Ruth, F.X. | | Deposit date: | 2013-03-07 | | Release date: | 2013-06-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel family of soluble minimal scaffolds provides structural insight into the catalytic domains of integral membrane metallopeptidases
J.Biol.Chem., 288, 2013
|
|
4JIU
 
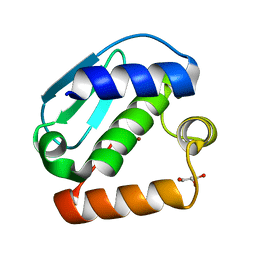 | | Crystal structure of the metallopeptidase zymogen of Pyrococcus abyssi abylysin | | Descriptor: | GLYCEROL, Proabylysin, ZINC ION | | Authors: | Lopez-Pelegrin, M, Cerda-Costa, N, Martinez-Jimenez, F, Cintas-Pedrola, A, Canals, A, Peinado, J.R, Marti-Renom, M.A, Lopez-Otin, C, Arolas, J.L, Gomis-Ruth, F.X. | | Deposit date: | 2013-03-07 | | Release date: | 2013-06-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A novel family of soluble minimal scaffolds provides structural insight into the catalytic domains of integral membrane metallopeptidases
J.Biol.Chem., 288, 2013
|
|
8RTH
 
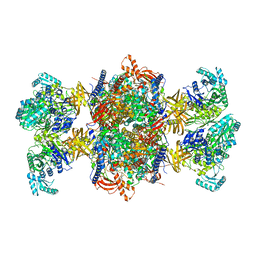 | | Trypanosoma brucei 3-methylcrotonyl-CoA carboxylase | | Descriptor: | 3-methylcrotonyl-CoA carboxylase, putative, 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ... | | Authors: | Ruiz, F.M, Plaza-Pegueroles, A, Fernandez-Tornero, C. | | Deposit date: | 2024-01-26 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | The cryo-EM structure of trypanosome 3-methylcrotonyl-CoA carboxylase provides mechanistic and dynamic insights into its enzymatic function.
Structure, 32, 2024
|
|
2FQ2
 
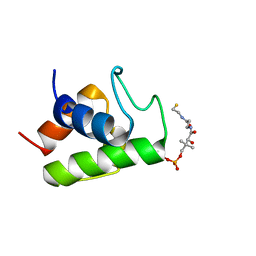 | | Solution structure of minor conformation of holo-acyl carrier protein from malaria parasite plasmodium falciparum | | Descriptor: | 4'-PHOSPHOPANTETHEINE, acyl carrier protein | | Authors: | Sharma, A.K, Sharma, S.K, Surolia, A, Surolia, N, Sarma, S.P. | | Deposit date: | 2006-01-17 | | Release date: | 2006-08-08 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structures of conformationally equilibrium forms of holo-acyl carrier protein (PfACP) from Plasmodium falciparum provides insight into the mechanism of activation of ACPs
Biochemistry, 45, 2006
|
|
5J51
 
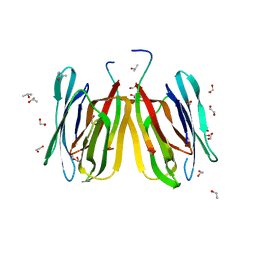 | | Structure of tetrameric jacalin complexed with Gal alpha-(1,4) Gal | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Surolia, A, Vijayan, M. | | Deposit date: | 2016-04-01 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Effect of linkage on the location of reducing and nonreducing sugars bound to jacalin.
IUBMB Life, 68, 2016
|
|
2FQ0
 
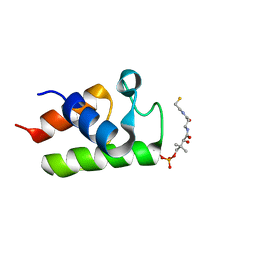 | | Solution structure of major conformation of holo-acyl carrier protein from malaria parasite plasmodium falciparum | | Descriptor: | 4'-PHOSPHOPANTETHEINE, acyl carrier protein | | Authors: | Sharma, A.K, Sharma, S.K, Surolia, A, Surolia, N, Sarma, S.P. | | Deposit date: | 2006-01-17 | | Release date: | 2006-08-08 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution structures of conformationally equilibrium forms of holo-acyl carrier protein (PfACP) from Plasmodium falciparum provides insight into the mechanism of activation of ACPs
Biochemistry, 45, 2006
|
|
3AZA
 
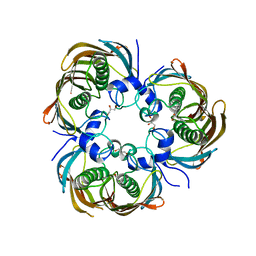 | | Beta-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Plasmodium falciparum in complex with NAS91-10 | | Descriptor: | 8-(benzyloxy)-5-chloroquinoline, Beta-hydroxyacyl-ACP dehydratase, GLYCEROL | | Authors: | Maity, K, Venkata, B.S, Kapoor, N, Surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2011-05-21 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the functional and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Plasmodium falciparum
J.Struct.Biol., 176, 2011
|
|
3AZ8
 
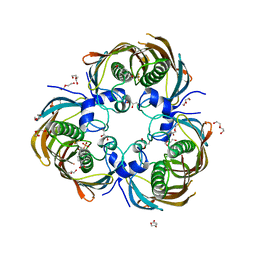 | | Beta-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Plasmodium falciparum in complex with NAS21 | | Descriptor: | 4,4,4-trifluoro-1-(4-nitrophenyl)butane-1,3-dione, Beta-hydroxyacyl-ACP dehydratase, CHLORIDE ION, ... | | Authors: | Maity, K, Venkata, B.S, Kapoor, N, Surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2011-05-20 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the functional and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Plasmodium falciparum
J.Struct.Biol., 176, 2011
|
|
3AZB
 
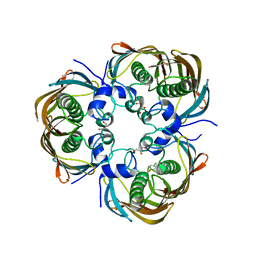 | | Beta-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Plasmodium falciparum in complex with NAS91-11 | | Descriptor: | 5-chloro-8-[(3-chlorobenzyl)oxy]quinoline, Beta-hydroxyacyl-ACP dehydratase, GLYCEROL | | Authors: | Maity, K, Venkata, B.S, Kapoor, N, Surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2011-05-21 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the functional and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Plasmodium falciparum
J.Struct.Biol., 176, 2011
|
|
3AZ9
 
 | | Beta-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Plasmodium falciparum in complex with NAS91 | | Descriptor: | 4-chloro-2-[(5-chloroquinolin-8-yl)oxy]phenol, Beta-hydroxyacyl-ACP dehydratase, GLYCEROL, ... | | Authors: | Maity, K, Venkata, B.S, Kapoor, N, Surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2011-05-21 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the functional and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Plasmodium falciparum
J.Struct.Biol., 176, 2011
|
|
6MR4
 
 | | Crystal structure of the Sth1 bromodomain from S.cerevisiae | | Descriptor: | Nuclear protein STH1/NPS1 | | Authors: | Seo, H.S, Hashimoto, H, Krolak, A, Debler, E.W, Blus, B.J. | | Deposit date: | 2018-10-11 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Substrate Affinity and Specificity of the ScSth1p Bromodomain Are Fine-Tuned for Versatile Histone Recognition.
Structure, 27, 2019
|
|
