4QSG
 
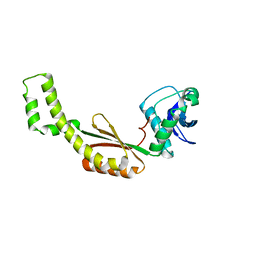 | | Crystal structure of gas vesicle protein GvpF from Microcystis aeruginosa | | Descriptor: | Gas vesicle protein | | Authors: | Xu, B.Y, Dai, Y.N, Zhou, K, Ren, Y.M, Liu, Y.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2014-07-03 | | Release date: | 2014-11-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the gas vesicle protein GvpF from the cyanobacterium Microcystis aeruginosa.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7JV7
 
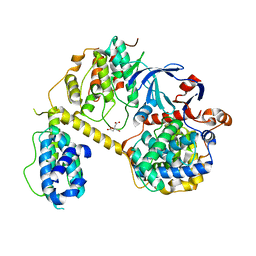 | | Crystal Structure of the yeast RNA Pol II CTD kinase CTDK-1 complex | | Descriptor: | CITRATE ANION, CTD kinase subunit alpha, CTD kinase subunit beta, ... | | Authors: | Xie, Y, Ren, Y. | | Deposit date: | 2020-08-20 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.850553 Å) | | Cite: | Structure and activation mechanism of the yeast RNA Pol II CTD kinase CTDK-1 complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KIR
 
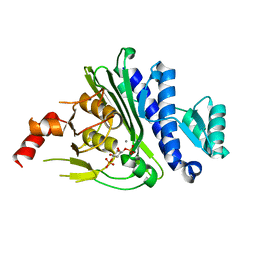 | | Crystal structure of inositol polyphosphate 1-phosphatase (INPP1) D54A mutant in complex with inositol (1,4)-bisphosphate | | Descriptor: | CALCIUM ION, D-MYO-INOSITOL-1,4-BISPHOSPHATE, Inositol polyphosphate 1-phosphatase | | Authors: | Dollins, D.E, Xiong, J.-P, Ren, Y, York, J.D. | | Deposit date: | 2020-10-24 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A structural basis for lithium and substrate binding of an inositide phosphatase.
J.Biol.Chem., 296, 2020
|
|
7KIO
 
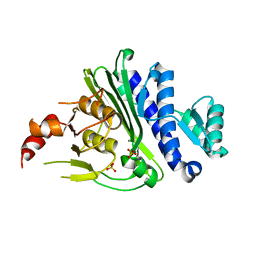 | | Crystal structure of inositol polyphosphate 1-phosphatase (INPP1) D54A mutant | | Descriptor: | CALCIUM ION, Inositol polyphosphate 1-phosphatase, SULFATE ION | | Authors: | Xiong, J.-P, Dollins, D.E, Ren, Y, York, J.D. | | Deposit date: | 2020-10-24 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A structural basis for lithium and substrate binding of an inositide phosphatase.
J.Biol.Chem., 296, 2020
|
|
4K6N
 
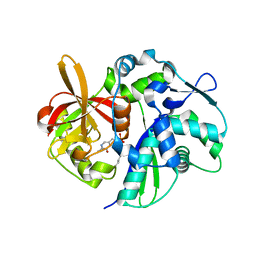 | | Crystal structure of yeast 4-amino-4-deoxychorismate lyase | | Descriptor: | Aminodeoxychorismate lyase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dai, Y.-N, Chi, C.-B, Zhou, K, Cheng, W, Jiang, Y.-L, Ren, Y.-M, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2013-04-16 | | Release date: | 2013-07-10 | | Last modified: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and catalytic mechanism of yeast 4-amino-4-deoxychorismate lyase
J.Biol.Chem., 288, 2013
|
|
7LUV
 
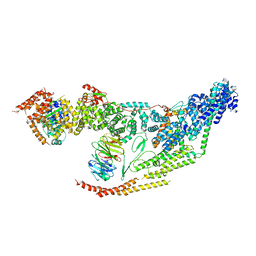 | | Cryo-EM structure of the yeast THO-Sub2 complex | | Descriptor: | ATP-dependent RNA helicase SUB2, THO complex subunit 2, THO complex subunit HPR1, ... | | Authors: | Xie, Y, Ren, Y. | | Deposit date: | 2021-02-23 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the yeast TREX complex and coordination with the SR-like protein Gbp2.
Elife, 10, 2021
|
|
7LEX
 
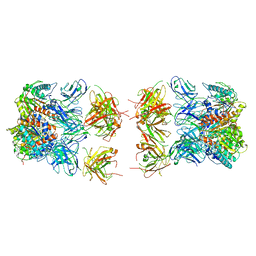 | |
7LF0
 
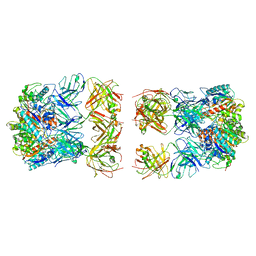 | |
7LEY
 
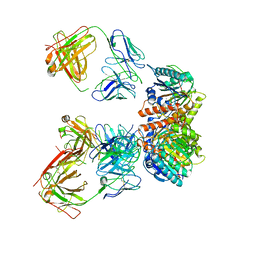 | | Trimeric human Arginase 1 in complex with mAb5 | | Descriptor: | Arginase-1, MANGANESE (II) ION, mAb5 heavy chain, ... | | Authors: | Gomez-Llorente, Y, Scapin, G, Palte, R.L. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Cryo-EM structures of inhibitory antibodies complexed with arginase 1 provide insight into mechanism of action.
Commun Biol, 4, 2021
|
|
6V8P
 
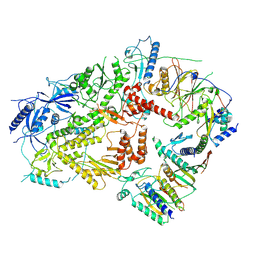 | | Structure of DNA Polymerase Zeta (Apo) | | Descriptor: | DNA polymerase delta small subunit, DNA polymerase delta subunit 3, DNA polymerase zeta catalytic subunit, ... | | Authors: | Malik, R, Gomez-Llorente, Y, Ubarretxena-Belandia, I, Aggarwal, A.K. | | Deposit date: | 2019-12-11 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure and mechanism of B-family DNA polymerase zeta specialized for translesion DNA synthesis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7LEZ
 
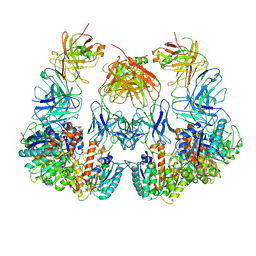 | | Trimeric human Arginase 1 in complex with mAb1 - 2 hArg:2 mAb1 complex | | Descriptor: | Arginase-1, MANGANESE (II) ION, mAb1 heavy chain, ... | | Authors: | Gomez-Llorente, Y, Scapin, G, Palte, R.L. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Cryo-EM structures of inhibitory antibodies complexed with arginase 1 provide insight into mechanism of action.
Commun Biol, 4, 2021
|
|
7LF2
 
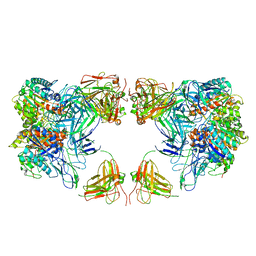 | | Trimeric human Arginase 1 in complex with mAb4 | | Descriptor: | Arginase-1, MANGANESE (II) ION, mAb4 monoclonal antibody heavy chain, ... | | Authors: | Gomez-Llorente, Y, Scapin, G, Palte, R.L. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Cryo-EM structures of inhibitory antibodies complexed with arginase 1 provide insight into mechanism of action.
Commun Biol, 4, 2021
|
|
7LF1
 
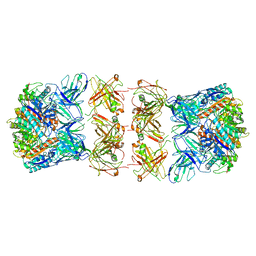 | | Trimeric human Arginase 1 in complex with mAb3 | | Descriptor: | Arginase-1, MANGANESE (II) ION, mAb3 heavy chain, ... | | Authors: | Gomez-Llorente, Y, Scapin, G, Palte, R.L. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Cryo-EM structures of inhibitory antibodies complexed with arginase 1 provide insight into mechanism of action.
Commun Biol, 4, 2021
|
|
6MRD
 
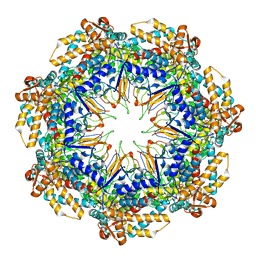 | | ADP-bound human mitochondrial Hsp60-Hsp10 half-football complex | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Gomez-Llorente, Y, Jebara, F, Patra, M, Malik, R, Nissemblat, S, Azem, A, Hirsch, J.A, Ubarretxena-Belandia, I. | | Deposit date: | 2018-10-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis for active single and double ring complexes in human mitochondrial Hsp60-Hsp10 chaperonin.
Nat Commun, 11, 2020
|
|
6MRC
 
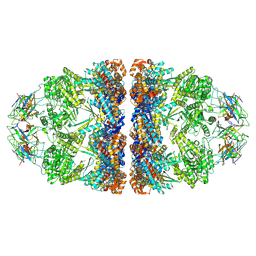 | | ADP-bound human mitochondrial Hsp60-Hsp10 football complex | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Gomez-Llorente, Y, Jebara, F, Patra, M, Malik, R, Nissemblat, S, Azem, A, Hirsch, J.A, Ubarretxena-Belandia, I. | | Deposit date: | 2018-10-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis for active single and double ring complexes in human mitochondrial Hsp60-Hsp10 chaperonin.
Nat Commun, 11, 2020
|
|
7MD4
 
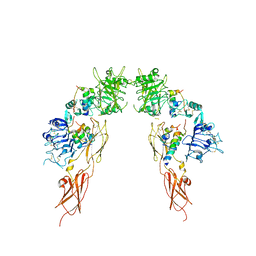 | | Insulin receptor ectodomain dimer complexed with two IRPA-3 partial agonists | | Descriptor: | Insulin B chain, Insulin chain A, Isoform Short of Insulin receptor, ... | | Authors: | Gomez-Llorente, Y, Zhou, H, Scapin, G. | | Deposit date: | 2021-04-03 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Functionally selective signaling and broad metabolic benefits by novel insulin receptor partial agonists.
Nat Commun, 13, 2022
|
|
8SK5
 
 | | Crystal structure of the SARS-CoV-2 neutralizing VHH 7A9 bound to the spike receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, anti-SARS-CoV-2 receptor binding domain VHH | | Authors: | Noland, C.L, Pande, K, Zhang, L, Zhou, H, Galli, J, Eddins, M, Gomez-Llorente, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Discovery and multimerization of cross-reactive single-domain antibodies against SARS-like viruses to enhance potency and address emerging SARS-CoV-2 variants.
Sci Rep, 13, 2023
|
|
7MD5
 
 | | Insulin receptor ectodomain dimer complexed with two IRPA-9 partial agonists | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gomez-Llorente, Y, Zhou, H, Scapin, G. | | Deposit date: | 2021-04-03 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Functionally selective signaling and broad metabolic benefits by novel insulin receptor partial agonists.
Nat Commun, 13, 2022
|
|
3E2N
 
 | |
3E2O
 
 | |
5EJX
 
 | | X-ray Free Electron Laser Structure of Cytochrome C Peroxidase | | Descriptor: | Cytochrome c peroxidase, mitochondrial, PHOSPHATE ION, ... | | Authors: | Doukov, T, Soltis, S.M, Baxter, E.L, Cohen, A, Song, J, McPhillips, S, Poulos, T.L, Meharenna, Y.T, Chreifi, G. | | Deposit date: | 2015-11-02 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the pristine peroxidase ferryl center and its relevance to proton-coupled electron transfer.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1SBR
 
 | | The structure and function of B. subtilis YkoF gene product: the complex with thiamin | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, CALCIUM ION, ykoF | | Authors: | Devedjiev, Y, Surendranath, Y, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2004-02-11 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure and Ligand Binding Properties of the B.subtilis YkoF Gene Product, a Member of a Novel Family of Thiamin/HMP-binding Proteins
J.Mol.Biol., 343, 2004
|
|
1S99
 
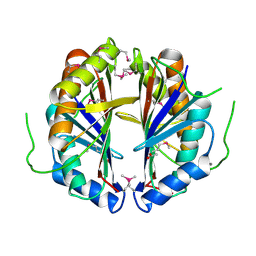 | | The structure and function of B. subtilis YkoF gene product: ligand free protein | | Descriptor: | ACETATE ION, CALCIUM ION, ykoF | | Authors: | Devedjiev, Y, Surendranath, Y, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2004-02-04 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Structure and Ligand Binding Properties of the B.subtilis YkoF Gene Product, a Member of a Novel Family of Thiamin/HMP-binding Proteins
J.Mol.Biol., 343, 2004
|
|
5EJT
 
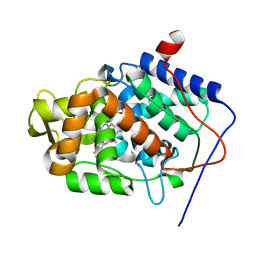 | | Thermally annealed ferryl Cytochrome C Peroxidase crystal structure | | Descriptor: | Cytochrome c peroxidase, mitochondrial, PHOSPHATE ION, ... | | Authors: | Doukov, T, Soltis, S.M, Baxter, E.L, Cohen, A, Song, J, McPhillips, S, Poulos, T.L, Meharenna, Y.T, Chreifi, G. | | Deposit date: | 2015-11-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the pristine peroxidase ferryl center and its relevance to proton-coupled electron transfer.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2BJO
 
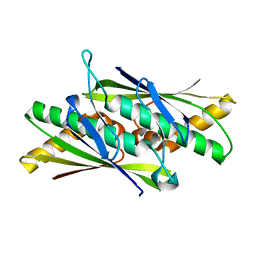 | | Crystal Structure of the Organic Hydroperoxide Resistance Protein OhrB of Bacillus subtilis | | Descriptor: | ORGANIC HYDROPEROXIDE RESISTANCE PROTEIN OHRB | | Authors: | Cooper, D.R, Surendranath, Y, Bielnicki, J, Devedjiev, Y, Joachimiak, A, Derewenda, Z.S. | | Deposit date: | 2005-02-04 | | Release date: | 2006-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Bacillus Subtilis Ohrb Hydroperoxide-Resistance Protein in a Fully Oxidized State.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
