2WKT
 
 | | BIOSYNTHETIC THIOLASE FROM Z. RAMIGERA. COMPLEX OF THE N316A MUTANT WITH COENZYME A. | | Descriptor: | ACETYL-COA ACETYLTRANSFERASE, CHLORIDE ION, COENZYME A, ... | | Authors: | Merilainen, G, Poikela, V, Kursula, P, Wierenga, R.K. | | Deposit date: | 2009-06-18 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Thiolase Reaction Mechanism: The Importance of Asn316 and His348 for Stabilizing the Enolate Intermediate of the Claisen Condensation.
Biochemistry, 48, 2009
|
|
1KQ2
 
 | | Crystal Structure of an Hfq-RNA Complex | | Descriptor: | 5'-R(*AP*UP*UP*UP*UP*UP*G)-3', Host factor for Q beta | | Authors: | Schumacher, M.A, Pearson, R.F, Moller, T, Valentin-Hansen, P, Brennan, R.G. | | Deposit date: | 2002-01-03 | | Release date: | 2002-07-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein.
EMBO J., 21, 2002
|
|
1SU5
 
 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-26 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1SW7
 
 | | Triosephosphate isomerase from Gallus gallus, loop 6 mutant K174N, T175S, A176S | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-30 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1KQ1
 
 | | 1.55 A Crystal structure of the pleiotropic translational regulator, Hfq | | Descriptor: | ACETIC ACID, Host Factor for Q beta | | Authors: | Schumacher, M.A, Pearson, R.F, Moller, T, Valentin-Hansen, P, Brennan, R.G. | | Deposit date: | 2002-01-03 | | Release date: | 2002-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein.
EMBO J., 21, 2002
|
|
1DKW
 
 | | CRYSTAL STRUCTURE OF TRIOSE-PHOSPHATE ISOMERASE WITH MODIFIED SUBSTRATE BINDING SITE | | Descriptor: | TERTIARY-BUTYL ALCOHOL, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Norledge, B.V, Lambeir, A.M, Abagyan, R.A, Rottman, A, Fernandez, A.M, Filimonov, V.V, Peter, M.G, Wierenga, R.K. | | Deposit date: | 1999-12-08 | | Release date: | 2000-11-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Modeling, mutagenesis, and structural studies on the fully conserved phosphate-binding loop (loop 8) of triosephosphate isomerase: toward a new substrate specificity.
Proteins, 42, 2001
|
|
2IB9
 
 | |
2IBY
 
 | |
2IB7
 
 | |
2WL5
 
 | | BIOSYNTHETIC THIOLASE FROM Z. RAMIGERA. COMPLEX OF THE H348N MUTANT WITH COENZYME A. | | Descriptor: | ACETYL-COA ACETYLTRANSFERASE, CHLORIDE ION, COENZYME A, ... | | Authors: | Merilainen, G, Poikela, V, Kursula, P, Wierenga, R.K. | | Deposit date: | 2009-06-22 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Thiolase Reaction Mechanism: The Importance of Asn316 and His348 for Stabilizing the Enolate Intermediate of the Claisen Condensation.
Biochemistry, 48, 2009
|
|
2WKV
 
 | | BIOSYNTHETIC THIOLASE FROM Z. RAMIGERA. COMPLEX OF THE N316D MUTANT WITH COENZYME A. | | Descriptor: | ACETYL-COA ACETYLTRANSFERASE, COENZYME A, SODIUM ION, ... | | Authors: | Merilainen, G, Poikela, V, Kursula, P, Wierenga, R.K. | | Deposit date: | 2009-06-18 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Thiolase Reaction Mechanism: The Importance of Asn316 and His348 for Stabilizing the Enolate Intermediate of the Claisen Condensation.
Biochemistry, 48, 2009
|
|
2WL4
 
 | | BIOSYNTHETIC THIOLASE FROM Z. RAMIGERA. COMPLEX OF THE H348A MUTANT WITH COENZYME A. | | Descriptor: | ACETYL-COA ACETYLTRANSFERASE, CHLORIDE ION, COENZYME A, ... | | Authors: | Merilainen, G, Poikela, V, Kursula, P, Wierenga, R.K. | | Deposit date: | 2009-06-22 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Thiolase Reaction Mechanism: The Importance of Asn316 and His348 for Stabilizing the Enolate Intermediate of the Claisen Condensation.
Biochemistry, 48, 2009
|
|
2WKU
 
 | | BIOSYNTHETIC THIOLASE FROM Z. RAMIGERA. THE N316H MUTANT. | | Descriptor: | ACETYL-COA ACETYLTRANSFERASE, D-mannose, SULFATE ION | | Authors: | Merilainen, G, Poikela, V, Kursula, P, Wierenga, R.K. | | Deposit date: | 2009-06-18 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Thiolase Reaction Mechanism: The Importance of Asn316 and His348 for Stabilizing the Enolate Intermediate of the Claisen Condensation.
Biochemistry, 48, 2009
|
|
1JUP
 
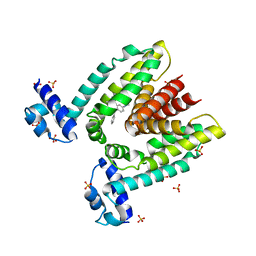 | | Crystal structure of the multidrug binding transcriptional repressor QacR bound to malachite green | | Descriptor: | HYPOTHETICAL TRANSCRIPTIONAL REGULATOR IN QACA 5'REGION, MALACHITE GREEN, SULFATE ION | | Authors: | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | Deposit date: | 2001-08-24 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural mechanisms of QacR induction and multidrug recognition.
Science, 294, 2001
|
|
1AW2
 
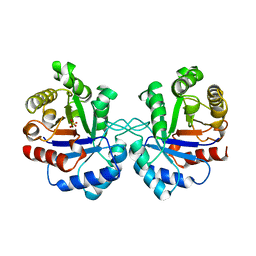 | | TRIOSEPHOSPHATE ISOMERASE OF VIBRIO MARINUS | | Descriptor: | SULFATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Maes, D, Zeelen, J.P, Wierenga, R.K. | | Deposit date: | 1997-10-09 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Triose-phosphate isomerase (TIM) of the psychrophilic bacterium Vibrio marinus. Kinetic and structural properties.
J.Biol.Chem., 273, 1998
|
|
1AW1
 
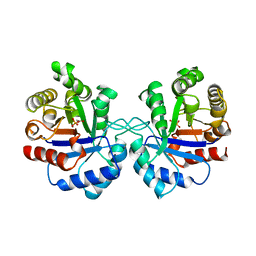 | |
5U56
 
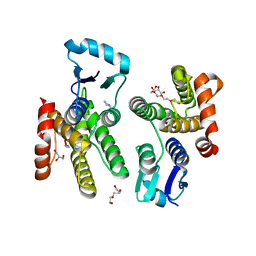 | | Structure of Francisella tularensis heterodimeric SspA (MglA-SspA) | | Descriptor: | GLYCEROL, Macrophage growth locus A, PENTAETHYLENE GLYCOL, ... | | Authors: | Cuthbert, B.J, Schumacher, M.A, Brennan, R.G. | | Deposit date: | 2016-12-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Dissection of the molecular circuitry controlling virulence in Francisella tularensis.
Genes Dev., 31, 2017
|
|
5UK7
 
 | | Escherichia coli Hfq bound to dsDNA | | Descriptor: | DNA (5'-D(P*CP*GP*GP*CP*AP*AP*AP*AP*AP*AP*CP*GP*GP*CP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*GP*CP*CP*GP*TP*TP*TP*TP*TP*TP*GP*CP*CP*G)-3'), RNA-binding protein Hfq, ... | | Authors: | Orans, J, Kovach, A.R, Brennan, R.G. | | Deposit date: | 2017-01-20 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Escherichia coli Hfq DNA complex reveals multifunctional nucleic acid binding site
To Be Published
|
|
1JUS
 
 | | Crystal structure of the multidrug binding transcriptional repressor QacR bound to rhodamine 6G | | Descriptor: | HYPOTHETICAL TRANSCRIPTIONAL REGULATOR IN QACA 5'REGION, RHODAMINE 6G, SULFATE ION | | Authors: | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | Deposit date: | 2001-08-27 | | Release date: | 2001-12-12 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural mechanisms of QacR induction and multidrug recognition.
Science, 294, 2001
|
|
1TJ7
 
 | | Structure determination and refinement at 2.44 A resolution of Argininosuccinate lyase from E. coli | | Descriptor: | Argininosuccinate lyase, GLYCEROL, PHOSPHATE ION | | Authors: | Bhaumik, P, Koski, M.K, Bergman, U, Wierenga, R.K. | | Deposit date: | 2004-06-03 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure determination and refinement at 2.44 A resolution of argininosuccinate lyase from Escherichia coli.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2A83
 
 | | Crystal structure of hla-b*2705 complexed with the glucagon receptor (gr) peptide (residues 412-420) | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Ruckert, C, Fiorillo, M.T, Loll, B, Moretti, R, Biesiadka, J, Saenger, W, Ziegler, A, Sorrentino, R, Uchanska-Ziegler, B. | | Deposit date: | 2005-07-07 | | Release date: | 2005-12-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational dimorphism of self-peptides and molecular mimicry in a disease-associated HLA-B27 subtype.
J.Biol.Chem., 281, 2006
|
|
1SSD
 
 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | SULFATE ION, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-24 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1IIG
 
 | | STRUCTURE OF TRYPANOSOMA BRUCEI BRUCEI TRIOSEPHOSPHATE ISOMERASE COMPLEXED WITH 3-PHOSPHONOPROPIONATE | | Descriptor: | 3-PHOSPHONOPROPANOIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Noble, M.E, Wierenga, R.K, Lambeir, A.M, Opperdoes, F.R, Thunnissen, A.M, Kalk, K.H, Groendijk, H, Hol, W.G.J. | | Deposit date: | 2001-04-23 | | Release date: | 2001-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The adaptability of the active site of trypanosomal triosephosphate isomerase as observed in the crystal structures of three different complexes.
Proteins, 10, 1991
|
|
1WN4
 
 | | NMR Structure of VoNTR | | Descriptor: | VoNTR protein | | Authors: | Dutton, J.L, Renda, R.F, Waine, C, Clark, R.J, Daly, N.L, Jennings, C.V, Anderson, M.A, Craik, D.J. | | Deposit date: | 2004-07-27 | | Release date: | 2004-09-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Conserved structural and sequence elements implicated in the processing of gene-encoded circular proteins
J.Biol.Chem., 279, 2004
|
|
1TTJ
 
 | |
