5HK0
 
 | |
6EVO
 
 | | Crystal structure the peptide-substrate-binding domain of human type II collagen prolyl 4-hydroxylase complexed with Pro-Pro-Gly-Pro-Arg-Gly-Pro-Pro-Gly. | | Descriptor: | DIMETHYL SULFOXIDE, PRO-PRO-GLY-PRO-ARG-GLY-PRO-PRO-GLY, Prolyl 4-hydroxylase subunit alpha-2, ... | | Authors: | Murthy, A.V, Sulu, R, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
Protein Sci., 27, 2018
|
|
6EVM
 
 | | Crystal structure of a Pro-9 complexed peptide-substrate-binding domain of human type II collagen prolyl 4-hydroxylase | | Descriptor: | DIMETHYL SULFOXIDE, Pro-9, Prolyl 4-hydroxylase subunit alpha-2, ... | | Authors: | Murthy, A.V, Sulu, R, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
Protein Sci., 27, 2018
|
|
6EVN
 
 | | Crystal structure of peptide-substrate-binding domain of human type II collagen prolyl 4-hydroxylase complex with Pro-Pro-Gly-Pro-Ala-Gly-Pro-Pro-Gly. | | Descriptor: | DIMETHYL SULFOXIDE, PRO-PRO-GLY-PRO-ALA-GLY-PRO-PRO-GLY, Prolyl 4-hydroxylase subunit alpha-2, ... | | Authors: | Murthy, A.V, Sulu, R, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
Protein Sci., 27, 2018
|
|
6EVL
 
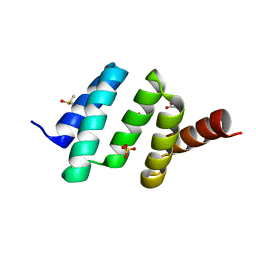 | | Crystal structure of an unlignaded peptide-substrate-binding domain of human type II collagen prolyl 4-hydroxylase | | Descriptor: | DIMETHYL SULFOXIDE, GLYCINE, Prolyl 4-hydroxylase subunit alpha-2, ... | | Authors: | Murthy, A.V, Sulu, R, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
Protein Sci., 27, 2018
|
|
6EVP
 
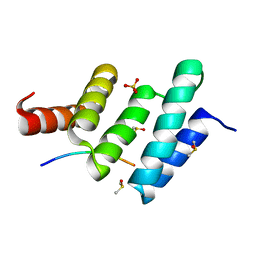 | | Crystal structure the peptide-substrate-binding domain of human type II collagen prolyl 4-hydroxylase complexed with Pro-Pro-Gly-Pro-Glu-Gly-Pro-Pro-Gly. | | Descriptor: | DIMETHYL SULFOXIDE, PRO-PRO-GLY-PRO-GLU-GLY-PRO-PRO-GLY, Prolyl 4-hydroxylase subunit alpha-2, ... | | Authors: | Murthy, A.V, Sulu, R, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
Protein Sci., 27, 2018
|
|
8FO1
 
 | |
7RLO
 
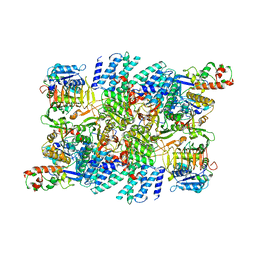 | | Structure of the human eukaryotic translation initiation factor 2B (eIF2B) in complex with a viral protein NSs | | Descriptor: | Non-structural protein NS-S, Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, ... | | Authors: | Wang, L, Schoof, M, Cogan, J, Lawrence, R, Boone, M, Wuerth, J, Frost, M, Walter, P. | | Deposit date: | 2021-07-26 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Viral evasion of the integrated stress response through antagonism of eIF2-P binding to eIF2B.
Nat Commun, 12, 2021
|
|
6TP5
 
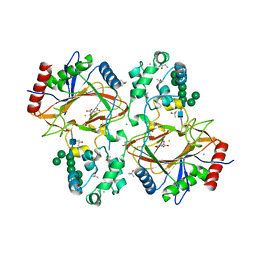 | | Crystal structure of human Transmembrane prolyl 4-hydroxylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Myllykoski, M, Sutinen, A, Koski, M.K, Kallio, J.P, Raasakka, A, Myllyharju, J, Wierenga, R.K, Koivunen, P. | | Deposit date: | 2019-12-12 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of transmembrane prolyl 4-hydroxylase reveals unique organization of EF and dioxygenase domains.
J.Biol.Chem., 296, 2020
|
|
8PD1
 
 | | Pseudomonas aeruginosa FabF C164A mutant in complex with N-isopropyl-1H-imidazole-4-carboxamide | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Georgiou, C, Brenk, R, Yadrykhinsky, V, Espeland, L.O. | | Deposit date: | 2023-06-11 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | New starting points for antibiotics targeting P. aeruginosa FabF discovered by crystallographic fragment screening followed by hit expansion
Chemrxiv, 2023
|
|
8PFZ
 
 | | Pseudomonas aeruginosa FabF C164A mutant in complex with(S)-2-(1H-pyrazole-3-carboxamido)butanoic acid | | Descriptor: | (2~{S})-2-(1~{H}-pyrazol-3-ylcarbonylamino)butanoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2, CHLORIDE ION, ... | | Authors: | Georgiou, C, Brenk, R, Yadrykhinsky, V, Espeland, L.O. | | Deposit date: | 2023-06-17 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | New starting points for antibiotics targeting P. aeruginosa FabF discovered by crystallographic fragment screening followed by hit expansion
Chemrxiv, 2023
|
|
8R1V
 
 | | Pseudomonas aeruginosa FabF C164A in complex with N-(1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazol-4-yl)-2-(4-methoxyphenoxy)acetamide | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Yadrykhinsky, V, Brenk, R. | | Deposit date: | 2023-11-02 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.087 Å) | | Cite: | Design, quality and validation of the EU-OPENSCREEN fragment library poised to a high-throughput screening collection.
Rsc Med Chem, 15, 2024
|
|
8R0I
 
 | | Pseudomonas aeruginosa FabF C164A in complex with 3-amino-N-(1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazol-4-yl)benzamide | | Descriptor: | 1,2-ETHANEDIOL, 3-azanyl-N-(1,5-dimethyl-3-oxidanylidene-2-phenyl-pyrazol-4-yl)benzamide, 3-oxoacyl-[acyl-carrier-protein] synthase 2, ... | | Authors: | Yadrykhinsky, V, Brenk, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Design, quality and validation of the EU-OPENSCREEN fragment library poised to a high-throughput screening collection.
Rsc Med Chem, 15, 2024
|
|
8QM1
 
 | | wild type Pa.FabF in complex cerulenin | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, 3-oxoacyl-[acyl-carrier-protein] synthase 2, ... | | Authors: | Georgiou, C, Brenk, R, Espeland, L.O. | | Deposit date: | 2023-09-21 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | New starting points for antibiotics targeting P. aeruginosa FabF discovered by crystallographic fragment screening followed by hit expansion
Chemrxiv, 2023
|
|
8QER
 
 | | Pseudomonas aeruginosa FabF C164A in complex with 4-(1H-pyrazole-3-carbonylamino)pentanoic acid | | Descriptor: | (4~{R})-4-(1~{H}-pyrazol-3-ylcarbonylamino)pentanoic acid, 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier-protein] synthase 2, ... | | Authors: | Yadrykhins'ky, V, Brenk, R, Georgiou, C. | | Deposit date: | 2023-09-01 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New starting points for antibiotics targeting P. aeruginosa FabF discovered by crystallographic fragment screening followed by hit expansion
Chemrxiv, 2023
|
|
8OQO
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-49 | | Descriptor: | GLYCEROL, Probable fatty oxidation protein FadB, Putative acyltransferase Rv0859, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8OQL
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-1 | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, GLYCEROL, Hexafluorophosphate anion, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8OQS
 
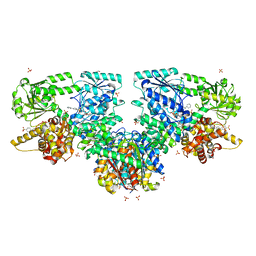 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-83 | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, 4-phenylbenzenesulfonic acid, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8OQR
 
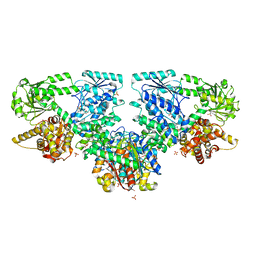 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-80 | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, 4-cyanobenzenesulfonic acid, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8OQN
 
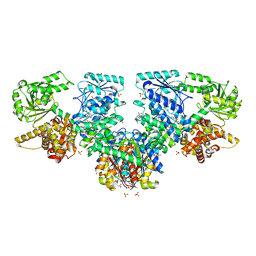 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-53 | | Descriptor: | 1-benzyl-1H-pyrazole-4-carboxylic acid, 3-hydroxyacyl-CoA dehydrogenase, Putative acyltransferase Rv0859, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8OQP
 
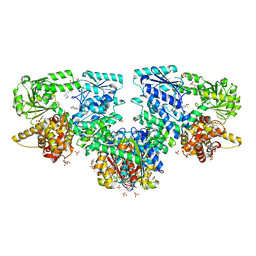 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-76 | | Descriptor: | 2-azanyl-5-sulfo-benzoic acid, 3-hydroxyacyl-CoA dehydrogenase, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8OQU
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-92 | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, 4-chloranylbenzenesulfonic acid, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8OQM
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-10 | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, 6-[(6-azanyl-4-oxidanyl-naphthalen-2-yl)sulfonylamino]-4-oxidanyl-naphthalene-2-sulfonic acid, 6-azanyl-4-oxidanyl-naphthalene-2-sulfonic acid, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8PF8
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-72 | | Descriptor: | (2~{R})-3-bis[2-methyl-5-(trifluoromethyl)pyrazol-3-yl]boranyloxypropane-1,2-diol, GLYCEROL, Probable fatty oxidation protein FadB, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-06-15 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7TRJ
 
 | | The eukaryotic translation initiation factor 2B from Homo sapiens with a H160D mutation in the beta subunit | | Descriptor: | Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, Translation initiation factor eIF-2B subunit delta, ... | | Authors: | Wang, L, Schoof, M, Lawrence, R, Boone, M, Frost, A, Walter, P. | | Deposit date: | 2022-01-29 | | Release date: | 2022-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A point mutation in the nucleotide exchange factor eIF2B constitutively activates the integrated stress response by allosteric modulation.
Elife, 11, 2022
|
|
