4YU8
 
 | | Crystal structure of Neuroblastoma suppressor of tumorigenicity 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Neuroblastoma suppressor of tumorigenicity 1 | | Authors: | Ren, J, Nettleship, J.E, Stammers, D.K, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2015-03-18 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Neuroblastoma suppressor of tumorigenicity 1
To Be Published
|
|
5C8C
 
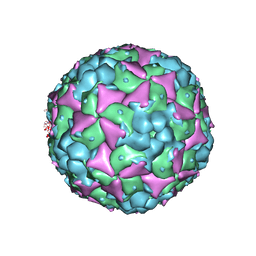 | | Crystal structure of recombinant coxsackievirus A16 capsid | | Descriptor: | CHLORIDE ION, POTASSIUM ION, STEARIC ACID, ... | | Authors: | Ren, J, Wang, X, Zhu, L, Hu, Z, Gao, Q, Yang, P, Li, X, Wang, J, Shen, X, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-06-25 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Coxsackievirus A16 Capsids with Native Antigenicity: Implications for Particle Expansion, Receptor Binding, and Immunogenicity.
J.Virol., 89, 2015
|
|
5GKK
 
 | |
1S1T
 
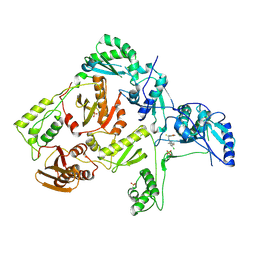 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with UC-781 | | Descriptor: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, PHOSPHATE ION, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
2BZ1
 
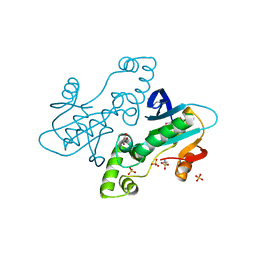 | | CRYSTAL STRUCTURE OF APO E. COLI GTP CYCLOHYDROLASE II | | Descriptor: | 2-AMINOETHANESULFONIC ACID, GLYCEROL, GTP CYCLOHYDROLASE II, ... | | Authors: | Ren, J, Kotaka, M, Lockyer, M, Lamb, H.K, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2005-08-09 | | Release date: | 2005-08-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | GTP Cyclohydrolase II Structure and Mechanism.
J.Biol.Chem., 280, 2005
|
|
1VRT
 
 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, MAGNESIUM ION | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-04-19 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
1VRU
 
 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | ALPHA-(2,6-DICHLOROPHENYL)-ALPHA-(2-ACETYL-5-METHYLANILINO)ACETAMIDE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-04-19 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
5C4W
 
 | | Crystal structure of coxsackievirus A16 | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SODIUM ION, ... | | Authors: | Ren, J, Wang, X, Zhu, L, Hu, Z, Gao, Q, Yang, P, Li, X, Wang, J, Shen, X, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-06-18 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of Coxsackievirus A16 Capsids with Native Antigenicity: Implications for Particle Expansion, Receptor Binding, and Immunogenicity.
J.Virol., 89, 2015
|
|
5C9A
 
 | | Crystal structure of empty coxsackievirus A16 particle | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SPHINGOSINE, ... | | Authors: | Ren, J, Wang, X, Zhu, L, Hu, Z, Gao, Q, Yang, P, Li, X, Wang, J, Shen, X, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-06-26 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of Coxsackievirus A16 Capsids with Native Antigenicity: Implications for Particle Expansion, Receptor Binding, and Immunogenicity.
J.Virol., 89, 2015
|
|
4MJS
 
 | | crystal structure of a PB1 complex | | Descriptor: | 1,2-ETHANEDIOL, Protein kinase C zeta type, Sequestosome-1 | | Authors: | Ren, J, Wang, Z.X, Wu, J.W. | | Deposit date: | 2013-09-04 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical insights into the homotypic PB1-PB1 complex between PKC zeta and p62
Sci China Life Sci, 57, 2014
|
|
1KLM
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH BHAP U-90152 | | Descriptor: | (1-(5-METHANSULPHONAMIDO-1H-INDOL-2-YL-CARBONYL)4-[METHYLAMINO)PYRIDINYL]PIPERAZINE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R.M, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1997-03-17 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Unique features in the structure of the complex between HIV-1 reverse transcriptase and the bis(heteroaryl)piperazine (BHAP) U-90152 explain resistance mutations for this nonnucleoside inhibitor.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
4IA0
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-bromo-2-{2-ethoxy-5-[(4-methylpiperazin-1-yl)sulfonyl]phenyl}-6-octylpyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ren, J, Chen, T, Xu, Y. | | Deposit date: | 2012-12-05 | | Release date: | 2014-01-01 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Exploration of the 5-bromopyrimidin-4(3H)-ones as potent inhibitors of PDE5.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1RT7
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH UC84 | | Descriptor: | 1-METHYL ETHYL 1-CHLORO-5-[[(5,6DIHYDRO-2-METHYL-1,4-OXATHIIN-3-YL)CARBONYL]AMINO]BENZOATE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1998-07-29 | | Release date: | 1999-07-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptase in complex with carboxanilide derivatives.
Biochemistry, 37, 1998
|
|
1RT6
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH UC38 | | Descriptor: | 1-METHYL ETHYL 2-CHLORO-5-[[[(1-METHYLETHOXY)THIOOXO]METHYL]AMINO]-BENZOATE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1998-07-29 | | Release date: | 1999-07-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptase in complex with carboxanilide derivatives.
Biochemistry, 37, 1998
|
|
1RT5
 
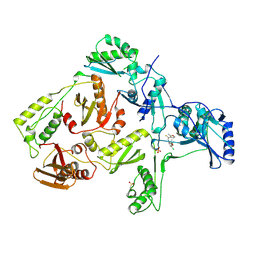 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH UC10 | | Descriptor: | HIV-1 REVERSE TRANSCRIPTASE, N-[4-CLORO-3-(T-BUTYLOXOME)PHENYL-2-METHYL-3-FURAN-CARBOTHIAMIDE, PHOSPHATE ION | | Authors: | Ren, J, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1998-07-29 | | Release date: | 1999-07-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptase in complex with carboxanilide derivatives.
Biochemistry, 37, 1998
|
|
1RT4
 
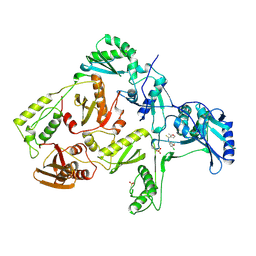 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH UC781 | | Descriptor: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1998-07-29 | | Release date: | 1999-07-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptase in complex with carboxanilide derivatives.
Biochemistry, 37, 1998
|
|
5DF8
 
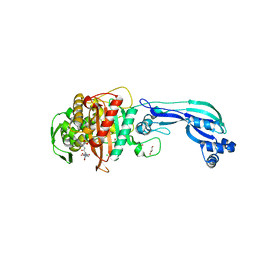 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH CEFOPERAZONE | | Descriptor: | (2R,4R,5R)-2-[(1R)-1-{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-(4-hydroxyphenyl)acetyl]amino}-2-oxoethyl]-5-methyl-1,3-thiazinane-4-carboxylic acid, CHLORIDE ION, Cell division protein, ... | | Authors: | Ren, J, Nettleship, J.E, Males, A, Stuart, D.I, Owens, R.J. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of penicillin-binding protein 3 in complexes with azlocillin and cefoperazone in both acylated and deacylated forms.
Febs Lett., 590, 2016
|
|
5DF7
 
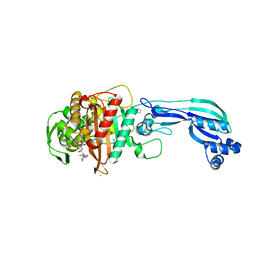 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH AZLOCILLIN | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-2-oxo-1-{[(2R)-2-{[(2-oxoimidazolidin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}ethyl]-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, Cell division protein, ... | | Authors: | Ren, J, Nettleship, J.E, Males, A, Stuart, D.I, Owens, R.J. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of penicillin-binding protein 3 in complexes with azlocillin and cefoperazone in both acylated and deacylated forms.
Febs Lett., 590, 2016
|
|
5DF9
 
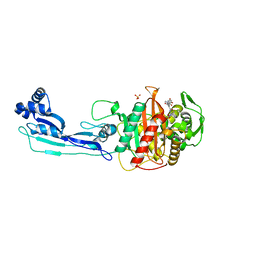 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 IN COMPLEX WITH DEACYLATED PRODUCT OF CEFOPERAZONE | | Descriptor: | (2R,5R)-2-[(R)-carboxy{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-(4-hydroxyphenyl)acetyl]amino}methyl]-5-methyl-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Cell division protein, GLYCEROL, ... | | Authors: | Ren, J, Nettleship, J.E, Males, A, Stuart, D.I, Owens, R.J. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of penicillin-binding protein 3 in complexes with azlocillin and cefoperazone in both acylated and deacylated forms.
Febs Lett., 590, 2016
|
|
4I9Z
 
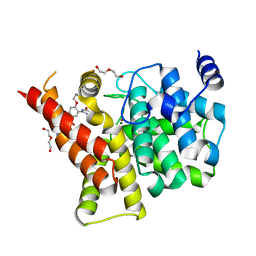 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-bromo-2-{5-[(4-methylpiperazin-1-yl)acetyl]-2-propoxyphenyl}-6-(propan-2-yl)pyrimidin-4(3H)-one, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Ren, J, Chen, T, Xu, Y. | | Deposit date: | 2012-12-05 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Exploration of the 5-bromopyrimidin-4(3H)-ones as potent inhibitors of PDE5.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4JGZ
 
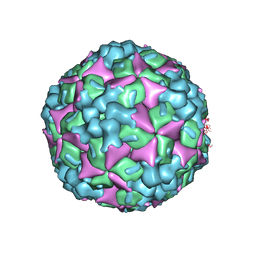 | | Crystal structure of human coxsackievirus A16 uncoating intermediate (space group I222) | | Descriptor: | Polyprotein, capsid protein VP1, capsid protein VP2, ... | | Authors: | Ren, J, Wang, X, Hu, Z, Gao, Q, Sun, Y, Li, X, Porta, C, Walter, T.S, Gilbert, R.J, Zhao, Y, Axford, D, Williams, M, McAuley, K, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Picornavirus uncoating intermediate captured in atomic detail.
Nat Commun, 4, 2013
|
|
4JGY
 
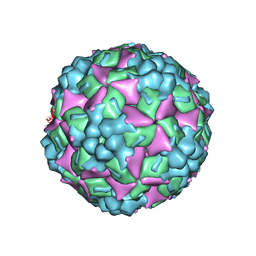 | | Crystal structure of human coxsackievirus A16 uncoating intermediate (space group P4232) | | Descriptor: | Polyprotein, capsid protein VP1, capsid protein VP2, ... | | Authors: | Ren, J, Wang, X, Hu, Z, Gao, Q, Sun, Y, Li, X, Porta, C, Walter, T.S, Gilbert, R.J, Zhao, Y, Axford, D, Williams, M, Mcauley, K, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Picornavirus uncoating intermediate captured in atomic detail.
Nat Commun, 4, 2013
|
|
1RT3
 
 | | AZT DRUG RESISTANT HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH 1051U91 | | Descriptor: | 6,11-DIHYDRO-11-ETHYL-6-METHYL-9-NITRO-5H-PYRIDO[2,3-B][1,5]BENZODIAZEPIN-5-ONE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1998-06-29 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 3'-Azido-3'-deoxythymidine drug resistance mutations in HIV-1 reverse transcriptase can induce long range conformational changes.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1RT1
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH MKC-442 | | Descriptor: | 6-BENZYL-1-ETHOXYMETHYL-5-ISOPROPYL URACIL, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Hopkins, A, Willcox, B, Jones, Y, Ross, C, Stammers, D, Stuart, D. | | Deposit date: | 1996-03-16 | | Release date: | 1997-04-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Complexes of HIV-1 reverse transcriptase with inhibitors of the HEPT series reveal conformational changes relevant to the design of potent non-nucleoside inhibitors.
J.Med.Chem., 39, 1996
|
|
1RT2
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH TNK-651 | | Descriptor: | 6-BENZYL-1-BENZYLOXYMETHYL-5-ISOPROPYL URACIL, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Hopkins, A, Willcox, B, Jones, Y, Ross, C, Stammers, D, Stuart, D. | | Deposit date: | 1996-03-16 | | Release date: | 1997-04-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Complexes of HIV-1 reverse transcriptase with inhibitors of the HEPT series reveal conformational changes relevant to the design of potent non-nucleoside inhibitors.
J.Med.Chem., 39, 1996
|
|
