6KH9
 
 | | Solution structure of bovine insulin amyloid intermediate-1 | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Ratha, B.N, Kar, R.K, Brender, J.B, Bhunia, A. | | Deposit date: | 2019-07-14 | | Release date: | 2020-08-12 | | Last modified: | 2020-11-18 | | Method: | SOLUTION NMR | | Cite: | High-resolution structure of a partially folded insulin aggregation intermediate.
Proteins, 88, 2020
|
|
6Z2P
 
 | | Crystal structure of catalytic inactive OgpA from Akkermansia muciniphila in complex with an O-glycopeptide (glycodrosocin) substrate | | Descriptor: | CALCIUM ION, Glycodrosocin, O-glycan protease, ... | | Authors: | Trastoy, B, Naegali, A, Anso, I, Sjogren, J, Guerin, M.E. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila.
Nat Commun, 11, 2020
|
|
6KHA
 
 | | Solution structure of bovine insulin amyloid intermediate-2 | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Ratha, B.N, Kar, R.K, Brender, J.B, Bhunia, A. | | Deposit date: | 2019-07-14 | | Release date: | 2020-08-12 | | Last modified: | 2020-11-18 | | Method: | SOLUTION NMR | | Cite: | High-resolution structure of a partially folded insulin aggregation intermediate.
Proteins, 88, 2020
|
|
1XAG
 
 | | CRYSTAL STRUCTURE OF STAPHLYOCOCCUS AUREUS 3-DEHYDROQUINATE SYNTHASE (DHQS) IN COMPLEX WITH ZN2+, NAD+ AND CARBAPHOSPHONATE | | Descriptor: | 3-dehydroquinate synthase, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Nichols, C.E, Ren, J, Leslie, K, Dhaliwal, B, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-08-25 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Comparison of ligand induced conformational changes and domain closure mechanisms, between prokaryotic and eukaryotic dehydroquinate synthases.
J.Mol.Biol., 343, 2004
|
|
1XAL
 
 | | CRYSTAL STRUCTURE OF STAPHLYOCOCCUS AUREUS 3-DEHYDROQUINATE SYNTHASE (DHQS) IN COMPLEX WITH ZN2+, NAD+ AND CARBAPHOSPHONATE (SOAK) | | Descriptor: | 3-dehydroquinate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION, ... | | Authors: | Nichols, C.E, Ren, J, Leslie, K, Dhaliwal, B, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-08-26 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparison of ligand induced conformational changes and domain closure mechanisms, between prokaryotic and eukaryotic dehydroquinate synthases.
J.Mol.Biol., 343, 2004
|
|
1XAJ
 
 | | CRYSTAL STRUCTURE OF STAPHLYOCOCCUS AUREUS 3-DEHYDROQUINATE SYNTHASE (DHQS) IN COMPLEX WITH ZN2+, NAD+ AND CARBAPHOSPHONATE | | Descriptor: | 3-dehydroquinate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION, ... | | Authors: | Nichols, C.E, Ren, J, Leslie, K, Dhaliwal, B, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-08-25 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Comparison of ligand induced conformational changes and domain closure mechanisms, between prokaryotic and eukaryotic dehydroquinate synthases.
J.Mol.Biol., 343, 2004
|
|
1GVG
 
 | | Crystal Structure of Clavaminate Synthase with Nitric Oxide | | Descriptor: | 2-OXOGLUTARIC ACID, CLAVAMINATE SYNTHASE 1, DEOXYGUANIDINOPROCLAVAMINIC ACID, ... | | Authors: | Zhang, Z.H, Ren, J, McKinnon, C.H, Clifton, I.J, Harlos, K, Schofield, C.J. | | Deposit date: | 2002-02-12 | | Release date: | 2003-02-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal Structure of a Clavaminate Synthase-Fe(II) -2-Oxoglutarate-Substrate-No Complex: Evidence for Metal Centered Rearrangements
FEBS Lett., 517, 2002
|
|
2BKA
 
 | | CC3(TIP30)Crystal Structure | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | El Omari, K, Bird, L.E, Nichols, C.E, Ren, J, Stammers, D.K. | | Deposit date: | 2005-02-14 | | Release date: | 2005-02-21 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Cc3 (Tip30): Implications for its Role as a Tumor Suppressor
J.Biol.Chem., 280, 2005
|
|
1XAH
 
 | | CRYSTAL STRUCTURE OF STAPHLYOCOCCUS AUREUS 3-DEHYDROQUINATE SYNTHASE (DHQS) IN COMPLEX WITH ZN2+ AND NAD+ | | Descriptor: | 3-dehydroquinate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Nichols, C.E, Ren, J, Leslie, K, Dhaliwal, B, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-08-25 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparison of ligand induced conformational changes and domain closure mechanisms, between prokaryotic and eukaryotic dehydroquinate synthases.
J.Mol.Biol., 343, 2004
|
|
1XAI
 
 | | CRYSTAL STRUCTURE OF STAPHLYOCOCCUS AUREUS 3-DEHYDROQUINATE SYNTHASE (DHQS) IN COMPLEX WITH ZN2+, NAD+ AND CARBAPHOSPHONATE | | Descriptor: | 3-dehydroquinate synthase, ZINC ION, [1R-(1ALPHA,3BETA,4ALPHA,5BETA)]-5-(PHOSPHONOMETHYL)-1,3,4-TRIHYDROXYCYCLOHEXANE-1-CARBOXYLIC ACID | | Authors: | Nichols, C.E, Ren, J, Leslie, K, Dhaliwal, B, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-08-25 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Comparison of ligand induced conformational changes and domain closure mechanisms, between prokaryotic and eukaryotic dehydroquinate synthases.
J.Mol.Biol., 343, 2004
|
|
1K6J
 
 | | Crystal structure of Nmra, a negative transcriptional regulator (Monoclinic form) | | Descriptor: | CHLORIDE ION, NmrA | | Authors: | Stammers, D.K, Ren, J, Leslie, K, Nichols, C.E, Lamb, H.K, Cocklin, S, Dodds, A, Hawkins, A.R. | | Deposit date: | 2001-10-16 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases.
EMBO J., 20, 2001
|
|
1K6I
 
 | | Crystal structure of Nmra, a negative transcriptional regulator (Trigonal form) | | Descriptor: | CHLORIDE ION, NmrA | | Authors: | Stammers, D.K, Ren, J, Leslie, K, Nichols, C.E, Lamb, H.K, Cocklin, S, Dodds, A, Hawkins, A.R. | | Deposit date: | 2001-10-16 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases.
EMBO J., 20, 2001
|
|
2CCK
 
 | | CRYSTAL STRUCTURE OF UNLIGANDED S. AUREUS THYMIDYLATE KINASE | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, THYMIDYLATE KINASE | | Authors: | Kotaka, M, Dhaliwal, B, Ren, J, Nichols, C.E, Angell, R, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2006-01-16 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structures of S. Aureus Thymidylate Kinase Reveal an Atypical Active Site Configuration and an Intermediate Conformational State Upon Substrate Binding
Protein Sci., 15, 2006
|
|
1K6X
 
 | | Crystal structure of Nmra, a negative transcriptional regulator in complex with NAD at 1.5 A resolution (Trigonal form) | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NmrA | | Authors: | Stammers, D.K, Ren, J, Leslie, K, Nichols, C.E, Lamb, H.K, Cocklin, S, Dodds, A, Hawkins, A.R. | | Deposit date: | 2001-10-17 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases.
EMBO J., 20, 2002
|
|
2CCJ
 
 | | Crystal structure of S. aureus thymidylate kinase complexed with thymidine monophosphate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, THYMIDINE-5'-PHOSPHATE, ... | | Authors: | Kotaka, M, Dhaliwal, B, Ren, J, Nichols, C.E, Angell, R, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2006-01-16 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of S. Aureus Thymidylate Kinase Reveal an Atypical Active Site Configuration and an Intermediate Conformational State Upon Substrate Binding
Protein Sci., 15, 2006
|
|
2Q5I
 
 | | Crystal structure of apo S581L Glycyl-tRNA synthetase mutant | | Descriptor: | Glycyl-tRNA synthetase, SULFATE ION | | Authors: | Cader, M.Z, Ren, J, James, P.A, Bird, L.E, Talbot, K, Stammers, D.K, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-06-01 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human wildtype and S581L-mutant glycyl-tRNA synthetase, an enzyme underlying distal spinal muscular atrophy.
Febs Lett., 581, 2007
|
|
2Q5H
 
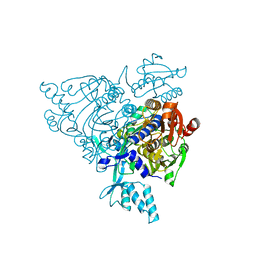 | | Crystal structure of apo-wildtype Glycyl-tRNA synthetase | | Descriptor: | Glycyl-tRNA synthetase | | Authors: | Cader, M.Z, Ren, J, James, P.A, Bird, L.E, Talbot, K, Stammers, D.K, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-06-01 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human wildtype and S581L-mutant glycyl-tRNA synthetase, an enzyme underlying distal spinal muscular atrophy.
Febs Lett., 581, 2007
|
|
6KH8
 
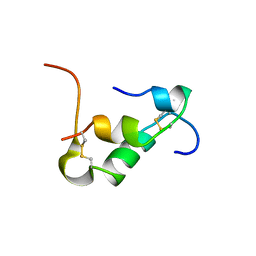 | | Solution structure of Zn free Bovine Pancreatic Insulin in 20% acetic acid-d4 (pH 1.9) | | Descriptor: | Insulin A Chain, Insulin B chain | | Authors: | Bhunia, A, Ratha, B.N, Kar, R.K, Brender, J.R. | | Deposit date: | 2019-07-14 | | Release date: | 2020-10-07 | | Last modified: | 2020-11-18 | | Method: | SOLUTION NMR | | Cite: | High-resolution structure of a partially folded insulin aggregation intermediate.
Proteins, 88, 2020
|
|
9FQ2
 
 | | Poliovirus 3C protease in H32 spacegroup | | Descriptor: | Protease 3C | | Authors: | Fairhead, M, Lithgo, R.M, MacLean, E.M, Bowesman-Jones, H, Aschenbrenner, J.C, Balcomb, B.H, Capkin, E, Chandran, A.V, Godoy, A.S, Marples, P.G, Fearon, D, von Delft, F, Koekemoer, L. | | Deposit date: | 2024-06-14 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Poliovirus 3C protease in H32 spacegroup
To Be Published
|
|
1DS0
 
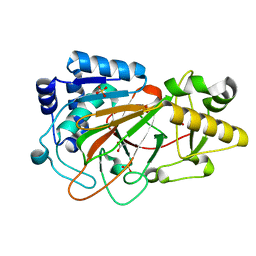 | | CRYSTAL STRUCTURE OF CLAVAMINATE SYNTHASE | | Descriptor: | ACETATE ION, CLAVAMINATE SYNTHASE 1, SULFATE ION | | Authors: | Zhang, Z.H, Ren, J, Stammers, D.K, Baldwin, J.E, Harlos, K, Schofield, C.J. | | Deposit date: | 2000-01-06 | | Release date: | 2000-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase.
Nat.Struct.Biol., 7, 2000
|
|
2CCG
 
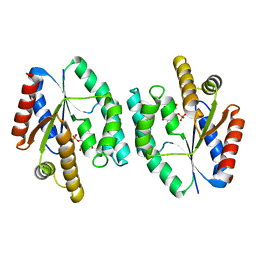 | | Crystal structure of His-tagged S. aureus thymidylate kinase complexed with thymidine monophosphate (TMP) | | Descriptor: | THYMIDINE-5'-PHOSPHATE, THYMIDYLATE KINASE | | Authors: | Kotaka, M, Dhaliwal, B, Ren, J, Nichols, C.E, Angell, R, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2006-01-16 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of S. Aureus Thymidylate Kinase Reveal an Atypical Active Site Configuration and an Intermediate Conformational State Upon Substrate Binding
Protein Sci., 15, 2006
|
|
1DRY
 
 | | CRYSTAL STRUCTURE OF CLAVAMINATE SYNTHASE IN COMPLEX WITH FE(II), 2-OXOGLUTARATE AND N-ALPHA-L-ACETYL ARGININE | | Descriptor: | 2-OXOGLUTARIC ACID, CLAVAMINATE SYNTHASE 1, FE (II) ION, ... | | Authors: | Zhang, Z.H, Ren, J, Stammers, D.K, Baldwin, J.E, Harlos, K, Schofield, C.J. | | Deposit date: | 2000-01-06 | | Release date: | 2000-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase.
Nat.Struct.Biol., 7, 2000
|
|
1DS1
 
 | | CRYSTAL STRUCTURE OF CLAVAMINATE SYNTHASE IN COMPLEX WITH FE(II) AND 2-OXOGLUTARATE | | Descriptor: | 2-OXOGLUTARIC ACID, CLAVAMINATE SYNTHASE 1, FE (II) ION, ... | | Authors: | Zhang, Z.H, Ren, J, Stammers, D.K, Baldwin, J.E, Harlos, K, Schofield, C.J. | | Deposit date: | 2000-01-06 | | Release date: | 2000-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase.
Nat.Struct.Biol., 7, 2000
|
|
1DRT
 
 | | CRYSTAL STRUCTURE OF CLAVAMINATE SYNTHASE IN COMPLEX WITH FE(II), 2-OXOGLUTARATE AND PROCLAVAMINIC ACID | | Descriptor: | 2-OXOGLUTARIC ACID, 5-AMINO-3-HYDROXY-2-(2-OXO-AZETIDIN-1-YL)-PENTANOIC ACID, CLAVAMINATE SYNTHASE 1, ... | | Authors: | Zhang, Z.H, Ren, J, Stammers, D.K, Baldwin, J.E, Harlos, K, Schofield, C.J. | | Deposit date: | 2000-01-06 | | Release date: | 2000-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase.
Nat.Struct.Biol., 7, 2000
|
|
2LCW
 
 | |
