4UU9
 
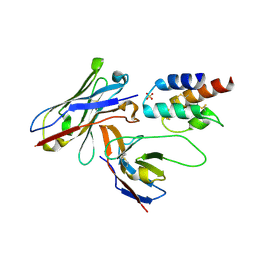 | | Crystal structure of the human c5a in complex with MEDI7814 a neutralising antibody | | Descriptor: | COMPLEMENT C5, MEDI7814, SULFATE ION | | Authors: | Colley, C, Sridharan, S, Dobson, C, Popovic, B, Debreczeni, J.E, Hargreaves, D, Edwards, B, Brennan, J, England, L, Fung, S, An Eghobamien, L, Sivars, U, Woods, R, Flavell, L, Renshaw, G.J, Wickson, K, Wilkinson, T, Davies, R, Bonnell, J, Warrener, P, Howes, R, Vaughan, T. | | Deposit date: | 2014-07-25 | | Release date: | 2015-08-12 | | Last modified: | 2019-02-27 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure and characterization of a high affinity C5a monoclonal antibody that blocks binding to C5aR1 and C5aR2 receptors.
MAbs, 10, 2018
|
|
4QVR
 
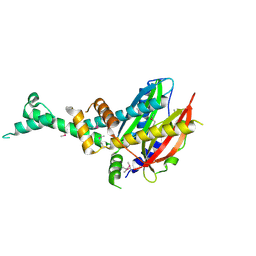 | | 2.3 Angstrom Crystal Structure of Hypothetical Protein FTT1539c from Francisella tularensis. | | Descriptor: | Uncharacterized hypothetical protein FTT_1539c | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Flores, K, Ren, G, Huntley, J.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-15 | | Release date: | 2014-07-30 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2.3 Angstrom Crystal Structure of Hypothetical Protein FTT1539c from Francisella tularensis.
TO BE PUBLISHED
|
|
1EHK
 
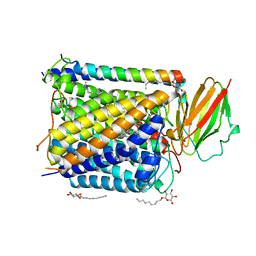 | | CRYSTAL STRUCTURE OF THE ABERRANT BA3-CYTOCHROME-C OXIDASE FROM THERMUS THERMOPHILUS | | Descriptor: | BA3-TYPE CYTOCHROME-C OXIDASE, COPPER (II) ION, DINUCLEAR COPPER ION, ... | | Authors: | Soulimane, T, Buse, G, Bourenkov, G.P, Bartunik, H.D, Huber, R, Than, M.E. | | Deposit date: | 2000-02-21 | | Release date: | 2001-02-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of the aberrant ba(3)-cytochrome c oxidase from thermus thermophilus.
EMBO J., 19, 2000
|
|
1EG5
 
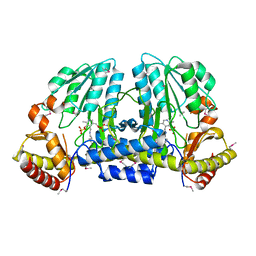 | | NIFS-LIKE PROTEIN | | Descriptor: | AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Kaiser, J.T, Clausen, T, Bourenkow, G.P, Bartunik, H.-D, Steinbacher, S, Huber, R. | | Deposit date: | 2000-02-13 | | Release date: | 2000-04-02 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a NifS-like protein from Thermotoga maritima: implications for iron sulphur cluster assembly.
J.Mol.Biol., 297, 2000
|
|
3CEI
 
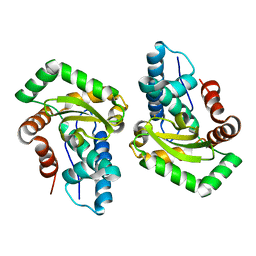 | | Crystal Structure of Superoxide Dismutase from Helicobacter pylori | | Descriptor: | FE (III) ION, SULFATE ION, Superoxide dismutase | | Authors: | Esposito, L, Seydel, A, Aiello, R, Sorrentino, G, Cendron, L, Zanotti, G, Zagari, A. | | Deposit date: | 2008-02-29 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the superoxide dismutase from Helicobacter pylori reveals a structured C-terminal extension
Biochim.Biophys.Acta, 1784, 2008
|
|
4UWM
 
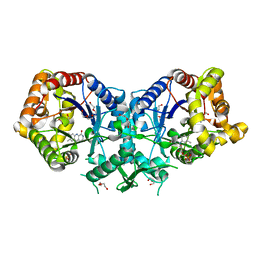 | | Type II Baeyer-Villiger monooxygenase.The oxygenating constituent of 3,6-diketocamphane monooxygenase from CAM plasmid of Pseudomonas putida in complex with FMN. | | Descriptor: | 3,6-DIKETOCAMPHANE 1,6 MONOOXYGENASE, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Isupov, M.N, Schroeder, E, Gibson, R.P, Beecher, J, Donadio, G, Saneei, V, Dcunha, S, McGhie, E.J, Sayer, C, Davenport, C.F, Lau, P.C, Hasegawa, Y, Iwaki, H, Kadow, M, Loschinski, K, Bornscheuer, U.T, Bourenkov, G, Littlechild, J.A. | | Deposit date: | 2014-08-12 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Oxygenating Constituent of 3,6-Diketocamphane Monooxygenase from the Cam Plasmid of Pseudomonas Putida: The First Crystal Structure of a Type II Baeyer-Villiger Monooxygenase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3D1I
 
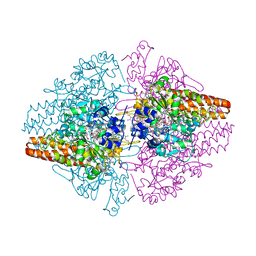 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with nitrite | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2008-05-06 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structural analysis of a novel octaheme cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens
J.Mol.Biol., 389, 2009
|
|
5A3Y
 
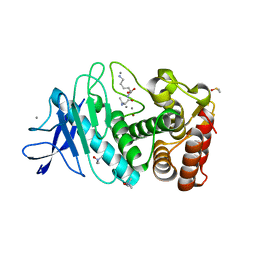 | | SAD structure of Thermolysin obtained by multi crystal data collection | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, LYSINE, ... | | Authors: | Zander, U, Bourenkov, G, Popov, A.N, de Sanctis, D, McCarthy, A.A, Svensson, O, Round, E.S, Gordeliy, V.I, Mueller-Dieckmann, C, Leonard, G.A. | | Deposit date: | 2015-06-04 | | Release date: | 2015-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Meshandcollect: An Automated Multi-Crystal Data-Collection Workflow for Synchrotron Macromolecular Crystallography Beamlines.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5A3Z
 
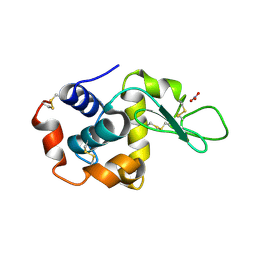 | | Structure of monoclinic Lysozyme obtained by multi crystal data collection | | Descriptor: | GLYCEROL, LYSOZYME, NITRATE ION | | Authors: | Zander, U, Bourenkov, G, Popov, A.N, de Sanctis, D, McCarthy, A.A, Svensson, O, Round, E.S, Gordeliy, V.I, Mueller-Dieckmann, C, Leonard, G.A. | | Deposit date: | 2015-06-04 | | Release date: | 2015-11-11 | | Last modified: | 2015-11-25 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Meshandcollect: An Automated Multi-Crystal Data-Collection Workflow for Synchrotron Macromolecular Crystallography Beamlines.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5A45
 
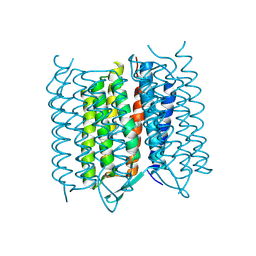 | | Structure of Bacteriorhodopsin obtained from 5um crystals by multi crystal data collection | | Descriptor: | BACTERIORHODOPSIN, RETINAL | | Authors: | Zander, U, Bourenkov, G, Popov, A.N, de Sanctis, D, McCarthy, A.A, Svensson, O, Round, E.S, Gordeliy, V.I, Mueller-Dieckmann, C, Leonard, G.A. | | Deposit date: | 2015-06-05 | | Release date: | 2015-11-11 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Meshandcollect: An Automated Multi-Crystal Data-Collection Workflow for Synchrotron Macromolecular Crystallography Beamlines.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5A47
 
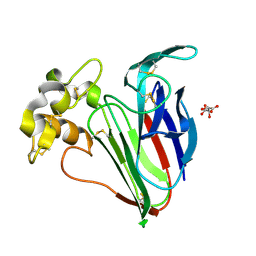 | | Structure of Thaumatin obtained by multi crystal data collection | | Descriptor: | L(+)-TARTARIC ACID, THAUMATIN-1 | | Authors: | Zander, U, Bourenkov, G, Popov, A.N, de Sanctis, D, McCarthy, A.A, Svensson, O, Round, E.S, Gordeliy, V.I, Mueller-Dieckmann, C, Leonard, G.A. | | Deposit date: | 2015-06-05 | | Release date: | 2015-11-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Meshandcollect: An Automated Multi-Crystal Data-Collection Workflow for Synchrotron Macromolecular Crystallography Beamlines.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6RX8
 
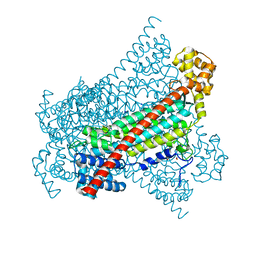 | |
3MJZ
 
 | | The crystal structure of native FG41 MSAD | | Descriptor: | FG41 Malonate Semialdehyde Decarboxylase | | Authors: | Guo, Y, Serrano, H, Poelarends, G.J, Johnson, W.H.Jr, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2010-04-13 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetic, Mutational, and Structural Analysis of Malonate Semialdehyde Decarboxylase from Coryneform Bacterium Strain FG41: Mechanistic Implications for the Decarboxylase and Hydratase Activities.
Biochemistry, 52, 2013
|
|
3LZZ
 
 | | Crystal structures of Cupin superfamily BbDUF985 from Branchiostoma belcheri tsingtauense in apo and GDP-bound forms | | Descriptor: | ACETATE ION, GUANOSINE-5'-DIPHOSPHATE, Putative uncharacterized protein | | Authors: | Du, Y, He, Y.-X, Saren, G, Zhang, X, Zhang, S.-C, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2010-03-02 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the apo and GDP-bound forms of a cupin-like protein BbDUF985 from Branchiostoma belcheri tsingtauense
Proteins, 2010
|
|
9BA5
 
 | |
9BA4
 
 | | Full-length cross-linked Contactin 2 (CNTN2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-2 | | Authors: | Liu, J.L, Fan, S.F, Ren, G.R, Rudenko, G.R. | | Deposit date: | 2024-04-03 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Molecular mechanism of Contactin 2 homophilic interaction
To Be Published
|
|
7SJR
 
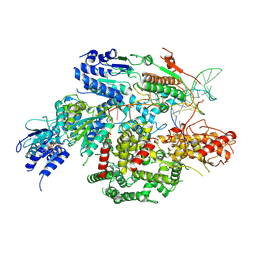 | | Cryo-EM structure of AdnA-AdnB(W325A) in complex with DNA and AMPPNP | | Descriptor: | DNA (70-MER), DNA helicase, IRON/SULFUR CLUSTER, ... | | Authors: | Wang, J, Warren, G.M, Shuman, S, Patel, D.J. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure-activity relationships at a nucleobase-stacking tryptophan required for chemomechanical coupling in the DNA resecting motor-nuclease AdnAB.
Nucleic Acids Res., 50, 2022
|
|
5LF0
 
 | | Human 20S proteasome complex with Epoxomicin at 2.4 Angstrom | | Descriptor: | CHLORIDE ION, EPOXOMICIN (peptide inhibitor), MAGNESIUM ION, ... | | Authors: | Schrader, J, Henneberg, F, Mata, R, Tittmann, K, Schneider, T.R, Stark, H, Bourenkov, G, Chari, A. | | Deposit date: | 2016-06-30 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The inhibition mechanism of human 20S proteasomes enables next-generation inhibitor design.
Science, 353, 2016
|
|
7QFC
 
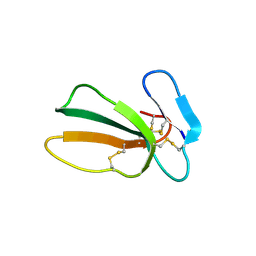 | | Crystal structure of cytotoxin 13 from Naja naja, orthorhombic form | | Descriptor: | Cytotoxin 13 | | Authors: | Samygina, V.R, Dubova, K.M, Bourenkov, G, Utkin, Y.N, Dubovskii, P.V. | | Deposit date: | 2021-12-05 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Variability in the Spatial Structure of the Central Loop in Cobra Cytotoxins Revealed by X-ray Analysis and Molecular Modeling.
Toxins, 14, 2022
|
|
7QHI
 
 | | Crystal structure of cytotoxin 13 from Naja naja, hexagonal form | | Descriptor: | Cytotoxin 13 | | Authors: | Samygina, V.R, Dubova, K.M, Bourenkov, G, Utkin, Y.N, Dubovskii, P.V. | | Deposit date: | 2021-12-12 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Variability in the Spatial Structure of the Central Loop in Cobra Cytotoxins Revealed by X-ray Analysis and Molecular Modeling.
Toxins, 14, 2022
|
|
7P76
 
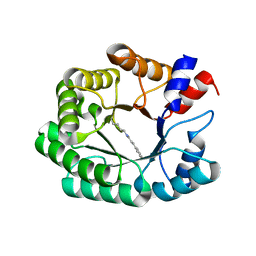 | | Re-engineered 2-deoxy-D-ribose-5-phosphate aldolase catalysing asymmetric Michael addition reactions, Schiff base complex with cinnamaldehyde | | Descriptor: | (2E)-3-phenylprop-2-enal, Deoxyribose-phosphate aldolase, GLYCEROL | | Authors: | Thunnissen, A.M.W.H, Rozeboom, H.J, Kunzendorf, A, Poelarends, G.J. | | Deposit date: | 2021-07-19 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unlocking Asymmetric Michael Additions in an Archetypical Class I Aldolase by Directed Evolution.
Acs Catalysis, 11, 2021
|
|
7PUO
 
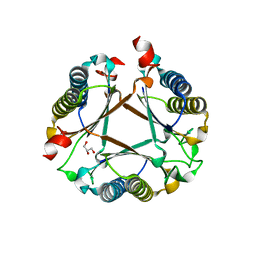 | | Structure of a fused 4-OT variant engineered for asymmetric Michael addition reactions | | Descriptor: | 2-hydroxymuconate tautomerase,Chains: A,B,C,D,E,F,2-hydroxymuconate tautomerase, CHLORIDE ION, GLYCEROL | | Authors: | Rozeboom, H.J, Thunnissen, A.M.W.H, Poelarends, G.J. | | Deposit date: | 2021-09-30 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Gene Fusion and Directed Evolution to Break Structural Symmetry and Boost Catalysis by an Oligomeric C-C Bond-Forming Enzyme.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7QUO
 
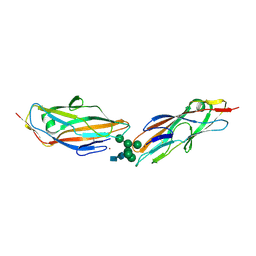 | | FimH lectin domain in complex with oligomannose-6 | | Descriptor: | FimH, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Bouckaert, J, Bourenkov, G.P. | | Deposit date: | 2022-01-18 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into a cooperative switch between one and two FimH bacterial adhesins binding pauci- and high-mannose type N-glycan receptors
J.Biol.Chem., 299, 2023
|
|
4RJK
 
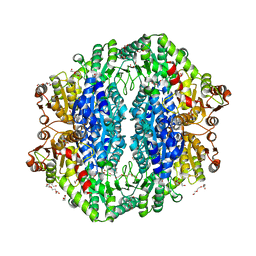 | | Acetolactate synthase from Bacillus subtilis bound to LThDP - crystal form II | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-CARBOXY-1-HYDROXYETHYL)-5-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, Acetolactate synthase, MAGNESIUM ION, ... | | Authors: | Sommer, B, von Moeller, H, Haack, M, Qoura, F, Langner, C, Bourenkov, G, Garbe, D, Brueck, T, Loll, B. | | Deposit date: | 2014-10-09 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detailed Structure-Function Correlations of Bacillus subtilis Acetolactate Synthase.
Chembiochem, 16, 2015
|
|
4RJJ
 
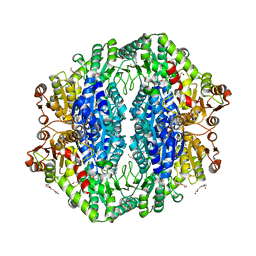 | | Acetolactate synthase from Bacillus subtilis bound to ThDP - crystal form II | | Descriptor: | ACETATE ION, Acetolactate synthase, MAGNESIUM ION, ... | | Authors: | Sommer, B, von Moeller, H, Haack, M, Qoura, F, Langner, C, Bourenkov, G, Garbe, D, Brueck, T, Loll, B. | | Deposit date: | 2014-10-09 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Detailed Structure-Function Correlations of Bacillus subtilis Acetolactate Synthase.
Chembiochem, 16, 2015
|
|
