5U0E
 
 | | Identification of a New Zinc Binding Chemotype by Fragment Screening | | Descriptor: | (5R)-5-benzyl-2-sulfanylidene-1,3-thiazolidin-4-one, 1,2-ETHANEDIOL, Carbonic anhydrase 2, ... | | Authors: | Peat, T.S, Poulsen, S.A, Ren, B, Dolezal, O, Woods, L.A, Mujumdar, P, Chrysanthopoulos, P.K. | | Deposit date: | 2016-11-23 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
5U0D
 
 | | Identification of a New Zinc Binding Chemotype by Fragment Screening | | Descriptor: | (5R)-5-(2,4-dimethoxyphenyl)-2-sulfanylidene-1,3-oxazolidin-4-one, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Peat, T.S, Poulsen, S.A, Ren, B, Dolezal, O, Woods, L.A, Mujumdar, P, Chrysanthopoulos, P.K. | | Deposit date: | 2016-11-23 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
5TY8
 
 | | Identification of a New Zinc Binding Chemotype by Fragment Screening | | Descriptor: | (5R)-5-phenyl-1,3-oxazolidine-2,4-dione, Carbonic anhydrase 2, ZINC ION | | Authors: | Peat, T.S, Poulsen, S.A, Ren, B, Dolezal, O, Woods, L.A, Mujumdar, P, Chrysanthopoulos, P.K. | | Deposit date: | 2016-11-18 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
5G47
 
 | | Structure of Gc glycoprotein from severe fever with thrombocytopenia syndrome virus in the trimeric postfusion conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, SFTSV GC | | Authors: | Halldorsson, S, Behrens, A.J, Harlos, K, Huiskonen, J.T, Elliott, R.M, Crispin, M, Brennan, B, Bowden, T.A. | | Deposit date: | 2016-05-05 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of a Phleboviral Envelope Glycoprotein Reveals a Consolidated Model of Membrane Fusion.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2RDF
 
 | |
1JFQ
 
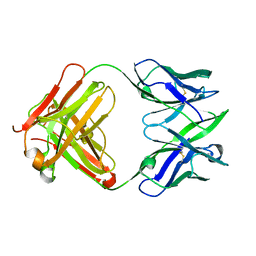 | | ANTIGEN-BINDING FRAGMENT OF THE MURINE ANTI-PHENYLARSONATE ANTIBODY 36-71, "FAB 36-71" | | Descriptor: | ANTIGEN-BINDING FRAGMENT OF ANTI-PHENYLARSONATE ANTIBODY | | Authors: | Parhami-Seren, B, Viswanathan, M, Strong, R.K, Margolies, M.N. | | Deposit date: | 2001-06-21 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of mutants of high-affinity and low-affinity
p-azophenylarsonate-specific antibodies generated by alanine
scanning of heavy chain
complementarity-determining region 2.
J.Immunol., 167, 2001
|
|
3HZX
 
 | |
3BDC
 
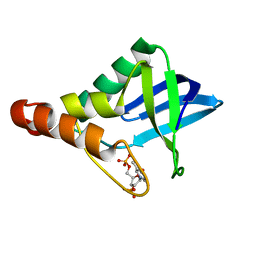 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Khangulov, V, Schlessman, J.L, Garcia-Moreno, B.E. | | Deposit date: | 2007-11-14 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular determinants of the pK(a) values of Asp and Glu residues in staphylococcal nuclease.
Proteins, 77, 2009
|
|
5FLU
 
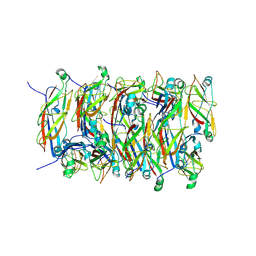 | | Structure of a Chaperone-Usher pilus reveals the molecular basis of rod uncoilin | | Descriptor: | PAP FIMBRIAL MAJOR PILIN PROTEIN | | Authors: | Hospenthal, M.K, Redzej, A, Dodson, K, Ukleja, M, Frenz, B, Hultgren, S.J, DiMaio, F, Egelman, E.H, Waksman, G. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of a Chaperone-Usher Pilus Reveals the Molecular Basis of Rod Uncoiling.
Cell(Cambridge,Mass.), 164, 2016
|
|
1U9R
 
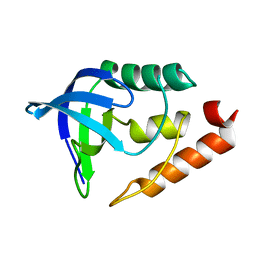 | |
1SNJ
 
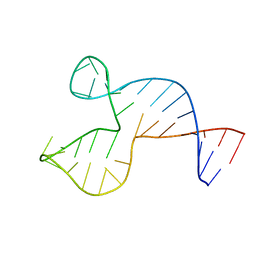 | | Solution structure of the DNA three-way junction with the A/C-stacked conformation | | Descriptor: | 36-MER | | Authors: | Wu, B, Girard, F, van Buuren, B, Schleucher, J, Tessari, M, Wijmenga, S. | | Deposit date: | 2004-03-11 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Global structure of a DNA three-way junction by solution NMR: towards prediction of 3H fold.
Nucleic Acids Res., 32, 2004
|
|
5V4S
 
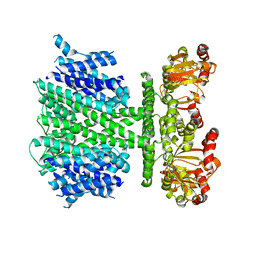 | | CryoEM Structure of a Prokaryotic Cyclic Nucleotide-Gated Ion Channel | | Descriptor: | Transporter, cation channel family / cyclic nucleotide-binding domain multi-domain protein | | Authors: | James, Z.M, Borst, A.J, Haitin, Y, Frenz, B, DiMaio, F, Zagotta, W.N, Veesler, D. | | Deposit date: | 2017-03-10 | | Release date: | 2017-04-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | CryoEM structure of a prokaryotic cyclic nucleotide-gated ion channel.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2QDB
 
 | |
4B5Q
 
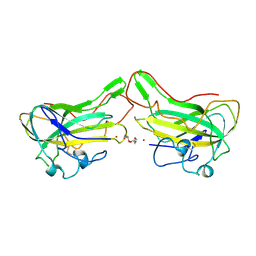 | | The lytic polysaccharide monooxygenase GH61D structure from the basidiomycota fungus Phanerochaete chrysosporium | | Descriptor: | COPPER (II) ION, GLYCEROL, GLYCOSIDE HYDROLASE FAMILY 61 PROTEIN D, ... | | Authors: | Wu, M, Beckham, G.T, Larsson, A.M, Ishida, T, Kim, S, Crowley, M.F, Payne, C.M, Horn, S.J, Westereng, B, Stahlberg, J, Eijsink, V.G.H, Sandgren, M. | | Deposit date: | 2012-08-07 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Computational Characterization of the Lytic Polysaccharide Monooxygenase Gh61D from the Basidiomycota Fungus Phanerochaete Chrysosporium
J.Biol.Chem., 288, 2013
|
|
2JJ9
 
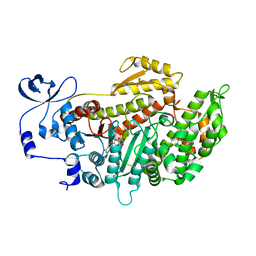 | | Crystal structure of myosin-2 in complex with ADP-metavanadate | | Descriptor: | ADP METAVANADATE, MAGNESIUM ION, MYOSIN-2 HEAVY CHAIN | | Authors: | Fedorov, R, Boehl, M, Tsiavaliaris, G, Hartmann, F.K, Baruch, P, Brenner, B, Martin, R, Knoelker, H.J, Gutzeit, H.O, Manstein, D.J. | | Deposit date: | 2008-03-25 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Mechanism of Pentabromopseudilin Inhibition of Myosin Motor Activity.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2ILT
 
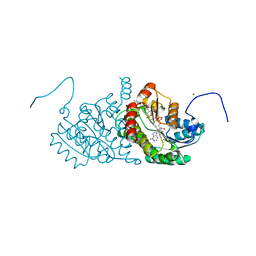 | | Human 11-beta-Hydroxysteroid Dehydrogenase (HSD1) with NADP and Adamantane Sulfone Inhibitor | | Descriptor: | 2-(2-CHLORO-4-FLUOROPHENOXY)-2-METHYL-N-[(1R,2S,3S,5S,7S)-5-(METHYLSULFONYL)-2-ADAMANTYL]PROPANAMIDE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Longenecker, K.L, Sorensen, B, Judge, R, Qin, W, Link, J.T. | | Deposit date: | 2006-10-03 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Adamantane sulfone and sulfonamide 11-beta-HSD1 Inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2HQW
 
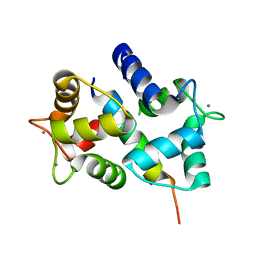 | | Crystal Structure of Ca2+/Calmodulin bound to NMDA Receptor NR1C1 peptide | | Descriptor: | CALCIUM ION, Calmodulin, Glutamate NMDA receptor subunit zeta 1 | | Authors: | Akyol, Z, Gakhar, L, Sorensen, B.R, Hell, J.H, Shea, M.A. | | Deposit date: | 2006-07-19 | | Release date: | 2007-11-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The NMDA Receptor NR1 C1 Region Bound to Calmodulin: Structural Insights into Functional Differences between Homologous Domains.
Structure, 15, 2007
|
|
2L4L
 
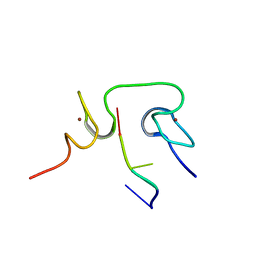 | | Structural insights into the cTAR DNA recognition by the HIV-1 Nucleocapsid protein: role of sugar deoxyriboses in the binding polarity of NC | | Descriptor: | 5'-D(*CP*TP*GP*G)-3', HIV-1 nucleocapsid protein NCp7, ZINC ION | | Authors: | Bazzi, A, Zargarian, L, Chaminade, F, Boudier, C, De Rocquigny, H, Rene, B, Mely, Y, Fosse, P, Mauffret, O. | | Deposit date: | 2010-10-08 | | Release date: | 2010-12-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the cTAR DNA recognition by the HIV-1 nucleocapsid protein: role of sugar deoxyriboses in the binding polarity of NC.
Nucleic Acids Res., 39, 2011
|
|
1EZN
 
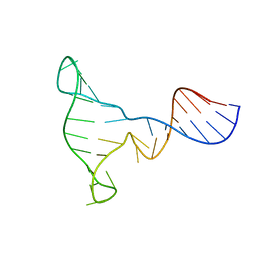 | | SOLUTION STRUCTURE OF A DNA THREE-WAY JUNCTION | | Descriptor: | DNA THREE-WAY JUNCTION | | Authors: | van Buuren, B.N.M, Overmars, F.J, Ippel, J.H, Altona, C, Wijmenga, S.S. | | Deposit date: | 2000-05-11 | | Release date: | 2001-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA three-way junction containing two unpaired thymidine bases. Identification of sequence features that decide conformer selection.
J.Mol.Biol., 304, 2000
|
|
2JHR
 
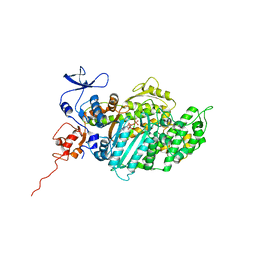 | | Crystal structure of myosin-2 motor domain in complex with ADP- metavanadate and pentabromopseudilin | | Descriptor: | ADP METAVANADATE, MAGNESIUM ION, MYOSIN-2 HEAVY CHAIN, ... | | Authors: | Fedorov, R, Boehl, M, Tsiavaliaris, G, Hartmann, F.K, Baruch, P, Brenner, B, Martin, R, Knoelker, H.J, Gutzeit, H.O, Manstein, D.J. | | Deposit date: | 2008-03-25 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Mechanism of Pentabromopseudilin Inhibition of Myosin Motor Activity.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2IZX
 
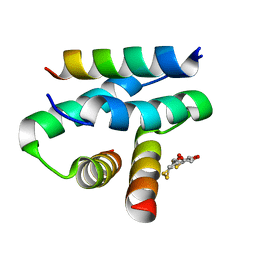 | | Molecular Basis of AKAP Specificity for PKA Regulatory Subunits | | Descriptor: | AKAP-IS, CAMP-DEPENDENT PROTEIN KINASE TYPE II-ALPHA REGULATORY SUBUNIT, DITHIANE DIOL | | Authors: | Gold, M.G, Lygren, B, Dokurno, P, Hoshi, N, McConnachie, G, Tasken, K, Carlson, C.R, Scott, J.D, Barford, D. | | Deposit date: | 2006-07-27 | | Release date: | 2006-11-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular Basis of Akap Specificity for Pka Regulatory Subunits.
Mol.Cell, 24, 2006
|
|
3EAB
 
 | | Crystal structure of Spastin MIT in complex with ESCRT III | | Descriptor: | CHMP1b, Spastin | | Authors: | Yang, D, Rimanchi, N, Renvoise, B, Lippincott-Schwartz, J, Blackstone, C, Hurley, J.H. | | Deposit date: | 2008-08-25 | | Release date: | 2008-11-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2IZY
 
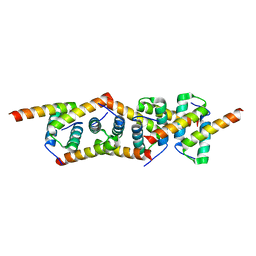 | | Molecular Basis of AKAP Specificity for PKA Regulatory Subunits | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE REGULATORY SUBUNIT II | | Authors: | Gold, M.G, Lygren, B, Dokurno, P, Hoshi, N, McConnachie, G, Tasken, K, Carlson, C.R, Scott, J.D, Barford, D. | | Deposit date: | 2006-07-27 | | Release date: | 2006-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Basis of Akap Specificity for Pka Regululatory Subunits
Mol.Cell, 24, 2006
|
|
5SZS
 
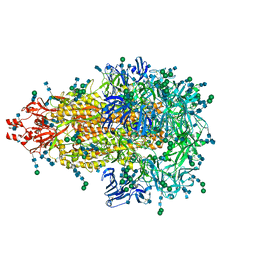 | | Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Walls, A.C, Tortorici, M.A, Frenz, B, Snijder, J, Li, W, Rey, F.A, DiMaio, F, Bosch, B.J, Veesler, D. | | Deposit date: | 2016-08-15 | | Release date: | 2016-09-14 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3JCL
 
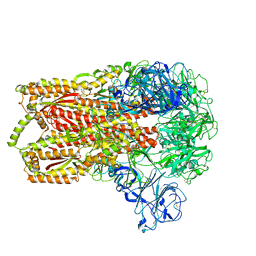 | | Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer | | Descriptor: | Spike glycoprotein | | Authors: | Walls, A.C, Tortorici, M.A, Bosch, B.J, Frenz, B, Rottier, P.J.M, DiMaio, F, Rey, F.A, Veesler, D. | | Deposit date: | 2015-12-21 | | Release date: | 2016-02-03 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer.
Nature, 531, 2016
|
|
