1MGX
 
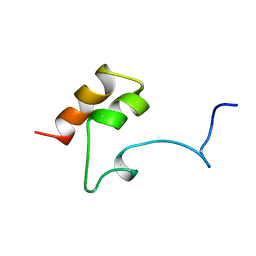 | | COAGULATION FACTOR, MG(II), NMR, 7 STRUCTURES (BACKBONE ATOMS ONLY) | | Descriptor: | COAGULATION FACTOR IX | | Authors: | Freedman, S.J, Furie, B.C, Furie, B, Baleja, J.D. | | Deposit date: | 1995-06-21 | | Release date: | 1996-11-08 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Identification of the phospholipid binding site in the vitamin K-dependent blood coagulation protein factor IX.
J.Biol.Chem., 271, 1996
|
|
1L3E
 
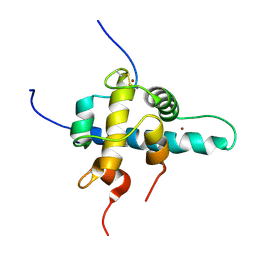 | | NMR Structures of the HIF-1alpha CTAD/p300 CH1 Complex | | Descriptor: | ZINC ION, hypoxia inducible factor-1 alpha subunit, p300 protein | | Authors: | Freedman, S.J, Sun, Z.J, Poy, F, Kung, A.L, Livingston, D.M, Wagner, G, Eck, M.J. | | Deposit date: | 2002-02-26 | | Release date: | 2002-04-24 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1NHL
 
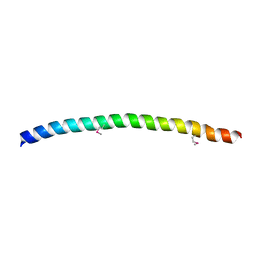 | | SNAP-23N Structure | | Descriptor: | Synaptosomal-associated protein 23 | | Authors: | Freedman, S.J, Song, H.K, Xu, Y, Eck, M.J. | | Deposit date: | 2002-12-19 | | Release date: | 2003-04-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Homotetrameric Structure of the SNAP-23 N-terminal Coiled-coil Domain
J.Biol.Chem., 278, 2003
|
|
1CFH
 
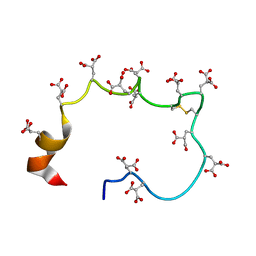 | | STRUCTURE OF THE METAL-FREE GAMMA-CARBOXYGLUTAMIC ACID-RICH MEMBRANE BINDING REGION OF FACTOR IX BY TWO-DIMENSIONAL NMR SPECTROSCOPY | | Descriptor: | COAGULATION FACTOR IX, FORMIC ACID | | Authors: | Freedman, S.J, Furie, B.C, Furie, B, Baleja, J.D. | | Deposit date: | 1995-02-26 | | Release date: | 1995-07-10 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the metal-free gamma-carboxyglutamic acid-rich membrane binding region of factor IX by two-dimensional NMR spectroscopy.
J.Biol.Chem., 270, 1995
|
|
1CFI
 
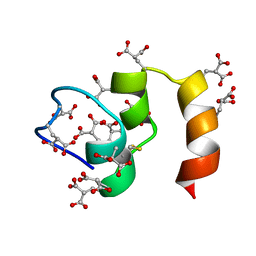 | |
1FSB
 
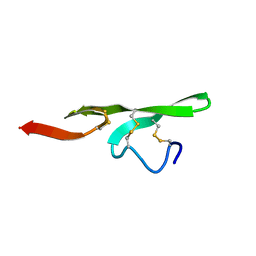 | | STRUCTURE OF THE EGF DOMAIN OF P-SELECTIN, NMR, 19 STRUCTURES | | Descriptor: | P-SELECTIN | | Authors: | Freedman, S.J, Sanford, D.G, Bachovchin, W.W, Furie, B.C, Baleja, J.D, Furie, B. | | Deposit date: | 1996-03-25 | | Release date: | 1997-04-01 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the epidermal growth factor domain of P-selectin.
Biochemistry, 35, 1996
|
|
1P4Q
 
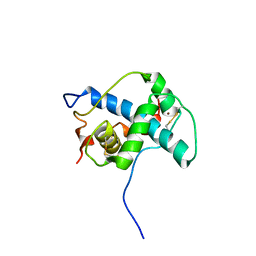 | | Solution structure of the CITED2 transactivation domain in complex with the p300 CH1 domain | | Descriptor: | Cbp/p300-interacting transactivator 2, E1A-associated protein p300, ZINC ION | | Authors: | Freedman, S.J, Sun, Z.-Y.J, Kung, A.L, France, D.S, Wagner, G, Eck, M.J. | | Deposit date: | 2003-04-23 | | Release date: | 2003-07-01 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural basis for negative regulation of hypoxia-inducible factor-1alpha by CITED2.
Nat.Struct.Biol., 10, 2003
|
|
5NYW
 
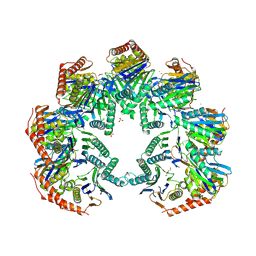 | | Anbu (ancestral beta-subunit) from Yersinia bercovieri | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, CHLORIDE ION, ... | | Authors: | Piasecka, A, Czapinska, H, Vielberg, M, Szczepanowski, R.H, Reed, S, Groll, M, Bochtler, M. | | Deposit date: | 2017-05-12 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | The Y. bercovieri Anbu crystal structure sheds light on the evolution of highly (pseudo)symmetric multimers.
J. Mol. Biol., 430, 2018
|
|
4ZJ1
 
 | | Crystal Structure of p-acrylamido-phenylalanine modified TEM1 beta-lactamase from Escherichia coli : V216AcrF mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase TEM, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Xiao, H, Nasertorabi, F, Choi, S, Han, G.W, Reed, S.A, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2015-04-28 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Exploring the potential impact of an expanded genetic code on protein function.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZJ2
 
 | | Crystal Structure of p-acrylamido-phenylalanine modified TEM1 beta-lactamase from Escherichia coli :E166N mutant | | Descriptor: | Beta-lactamase TEM | | Authors: | Xiao, H, Nasertorabi, F, Choi, S, Han, G.W, Reed, S.A, Stevens, C.S, Schultz, P.G. | | Deposit date: | 2015-04-28 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploring the potential impact of an expanded genetic code on protein function.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZJ3
 
 | | Crystal structure of cephalexin bound acyl-enzyme intermediate of Val216AcrF mutant TEM1 beta-lactamase from Escherichia coli: E166N and V216AcrF mutant. | | Descriptor: | Beta-lactamase TEM | | Authors: | Xiao, H, Nasertorabi, F, Choi, S, Han, G.W, Reed, S.A, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exploring the potential impact of an expanded genetic code on protein function.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4CCQ
 
 | | Crystal structure of the thioredoxin reductase from Entamoeba histolytica with NADP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Parsonage, D, Kells, P.M, Hirata, K, Debnath, A, Poole, L.B, McKerrow, J.H, Reed, S.L, Podust, L.M. | | Deposit date: | 2013-10-24 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-Ray Structures of Thioredoxin and Thioredoxin Reductase from Entamoeba Histolytica and Prevailing Hypothesis of the Mechanism of Auranofin Action.
J.Struct.Biol., 194, 2016
|
|
4CW9
 
 | | Entamoeba histolytica thiredoxin C34S mutant | | Descriptor: | THIOREDOXIN | | Authors: | Parsonage, D, Kells, P.M, Vieira, D.F, Hirata, K, Debnath, A, Poole, L.B, McKerrow, J.H, Reed, S.L, Podust, L.M. | | Deposit date: | 2014-04-01 | | Release date: | 2015-05-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | X-Ray Structures of Thioredoxin and Thioredoxin Reductase from Entamoeba Histolytica and Prevailing Hypothesis of the Mechanism of Auranofin Action.
J.Struct.Biol., 194, 2016
|
|
4CCR
 
 | | Crystal structure of the thioredoxin reductase apoenzyme from Entamoeba histolytica in the absence of the NADP cofactor | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GOLD ION, THIOREDOXIN REDUCTASE | | Authors: | Parsonage, D, Kells, P.M, Hirata, K, Debnath, A, Poole, L.B, McKerrow, J.H, Reed, S.L, Podust, L.M. | | Deposit date: | 2013-10-25 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | X-Ray Structures of Thioredoxin and Thioredoxin Reductase from Entamoeba Histolytica and Prevailing Hypothesis of the Mechanism of Auranofin Action.
J.Struct.Biol., 194, 2016
|
|
4CBQ
 
 | | Crystal structure of the thioredoxin reductase from Entamoeba histolytica with auranofin Au(I) bound to Cys286 | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GOLD ION, ... | | Authors: | Parsonage, D, Kells, P.M, Hirata, K, Debnath, A, Poole, L.B, McKerrow, J.H, Reed, S.L, Podust, L.M. | | Deposit date: | 2013-10-16 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | X-Ray Structures of Thioredoxin and Thioredoxin Reductase from Entamoeba Histolytica and Prevailing Hypothesis of the Mechanism of Auranofin Action.
J.Struct.Biol., 194, 2016
|
|
1QB3
 
 | | CRYSTAL STRUCTURE OF THE CELL CYCLE REGULATORY PROTEIN CKS1 | | Descriptor: | CYCLIN-DEPENDENT KINASES REGULATORY SUBUNIT | | Authors: | Bourne, Y, Watson, M.H, Arvai, A.S, Bernstein, S.L, Reed, S.I, Tainer, J.A. | | Deposit date: | 1999-04-30 | | Release date: | 2000-08-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure and mutational analysis of the Saccharomyces cerevisiae cell cycle regulatory protein Cks1: implications for domain swapping, anion binding and protein interactions.
Structure Fold.Des., 8, 2000
|
|
7XNZ
 
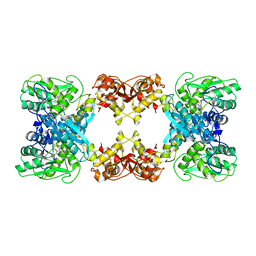 | | Native cystathionine beta-synthase of Mycobacterium tuberculosis. | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative cystathionine beta-synthase Rv1077 | | Authors: | Bandyopadhyay, P, Pramanick, I, Biswas, R, Sabarinath, P.S, Sreedharan, S, Singh, S, Rajmani, R, Laxman, S, Dutta, S, Singh, A. | | Deposit date: | 2022-04-30 | | Release date: | 2022-05-25 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | S-Adenosylmethionine-responsive cystathionine beta-synthase modulates sulfur metabolism and redox balance in Mycobacterium tuberculosis.
Sci Adv, 8, 2022
|
|
7XOH
 
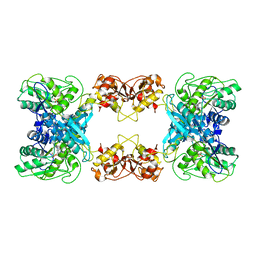 | | Cystathionine beta-synthase of Mycobacterium tuberculosis in the presence of S-adenosylmethionine. | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative cystathionine beta-synthase Rv1077 | | Authors: | Bandyopadhyay, P, Pramanick, I, Biswas, R, Sabarinath, P.S, Sreedharan, S, Singh, S, Rajmani, R, Laxman, S, Dutta, S, Singh, A. | | Deposit date: | 2022-05-01 | | Release date: | 2022-05-25 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | S-Adenosylmethionine-responsive cystathionine beta-synthase modulates sulfur metabolism and redox balance in Mycobacterium tuberculosis.
Sci Adv, 8, 2022
|
|
7XOY
 
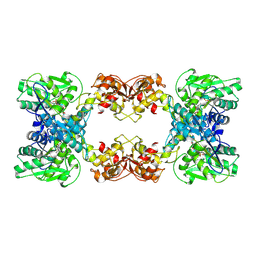 | | Cystathionine beta-synthase of Mycobacterium tuberculosis in the presence of S-adenosylmethionine and serine. | | Descriptor: | Putative cystathionine beta-synthase Rv1077, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-SERINE | | Authors: | Bandyopadhyay, P, Pramanick, I, Biswas, R, Sabarinath, P.S, Sreedharan, S, Singh, S, Rajmani, R, Laxman, S, Dutta, S, Singh, A. | | Deposit date: | 2022-05-01 | | Release date: | 2022-05-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | S-Adenosylmethionine-responsive cystathionine beta-synthase modulates sulfur metabolism and redox balance in Mycobacterium tuberculosis.
Sci Adv, 8, 2022
|
|
