3K4D
 
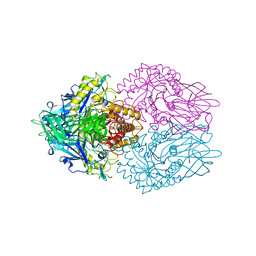 | | Crystal structure of E. coli beta-glucuronidase with the glucaro-d-lactam inhibitor bound | | Descriptor: | (2S,3R,4S,5R)-3,4,5-trihydroxy-6-oxopiperidine-2-carboxylic acid, Beta-glucuronidase | | Authors: | Wallace, B.D, Redinbo, M.R. | | Deposit date: | 2009-10-05 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Alleviating cancer drug toxicity by inhibiting a bacterial enzyme.
Science, 330, 2010
|
|
3K9B
 
 | |
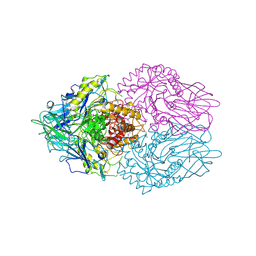
3K4A
 
 | |
1M13
 
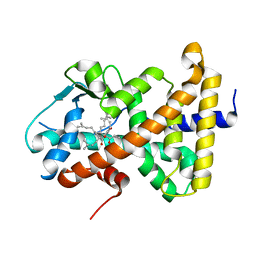 | | Crystal Structure of the Human Pregane X Receptor Ligand Binding Domain in Complex with Hyperforin, a Constituent of St. John's Wort | | Descriptor: | 4-HYDROXY-5-ISOBUTYRYL-6-METHYL-1,3,7-TRIS-(3-METHYL-BUT-2-ENYL)-6-(4-METHYL-PENT-3-ENYL)-BICYCLO[3.3.1]NON-3-ENE-2,9-DIONE, Orphan Nuclear Receptor PXR | | Authors: | Watkins, R.E, Maglich, J.M, Moore, L.B, Wisely, G.B, Noble, S.M, Davis-Searles, P.R, Lambert, M.H, Kliewer, S.A, Redinbo, M.R. | | Deposit date: | 2002-06-17 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.1 A Crystal Structure of Human PXR in Complex with the St. John's Wort Compound Hyperforin
Biochemistry, 42, 2003
|
|
1MX1
 
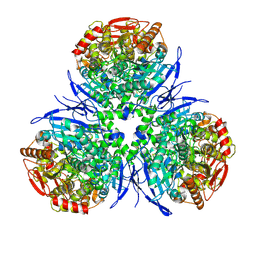 | | Crystal Structure of Human Liver Carboxylesterase in complex with tacrine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Bencharit, S, Morton, C.L, Hyatt, J.L, Kuhn, P, Danks, M.K, Potter, P.M, Redinbo, M.R. | | Deposit date: | 2002-10-01 | | Release date: | 2003-04-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Human Carboxylesterase 1 Complexed with the Alzheimer's
Drug Tacrine: From Binding Promiscuity to Selective Inhibition
CHEM.BIOL., 10, 2003
|
|
1NRL
 
 | | Crystal Structure of the human PXR-LBD in complex with an SRC-1 coactivator peptide and SR12813 | | Descriptor: | Nuclear Receptor Coactivator 1 isoform 3, Orphan nuclear receptor PXR, [2-(3,5-DI-TERT-BUTYL-4-HYDROXY-PHENYL)-1-(DIETHOXY-PHOSPHORYL)-VINYL]-PHOSPHONIC ACID DIETHLYL ESTER | | Authors: | Watkins, R.E, Davis-Searles, P.R, Lambert, M.H, Redinbo, M.R. | | Deposit date: | 2003-01-25 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Coactivator binding promotes the specific interaction between ligand and the pregnane X receptor
J.Mol.Biol., 331, 2003
|
|
1MX5
 
 | | Crystal Structure of Human Liver Carboxylesterase in complexed with homatropine, a cocaine analogue | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, HOMOTROPINE, ... | | Authors: | Bencharit, S, Morton, C.L, Xue, Y, Potter, P.M, Redinbo, M.R. | | Deposit date: | 2002-10-01 | | Release date: | 2003-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Heroin and Cocaine Metabolism by a Promiscuous Human Drug-Processing Enzyme
Nat.Struct.Biol., 10, 2003
|
|
1MX9
 
 | | Crystal Structure of Human Liver Carboxylesterase in complexed with naloxone methiodide, a heroin analogue | | Descriptor: | (5A,17R)-4,5-EPOXY-3,14-DIHYDROXY-17-METHYL-6-OXO-17-(2-PROPENYL)-MORPHINANIUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, liver Carboxylesterase I | | Authors: | Bencharit, S, Morton, C.L, Xue, Y, Potter, P.M, Redinbo, M.R. | | Deposit date: | 2002-10-01 | | Release date: | 2003-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis of Heroin and Cocaine Metabolism by a Promiscuous Human Drug-Processing Enzyme
Nat.Struct.Biol., 10, 2003
|
|
4EQW
 
 | | Crystal Structure of the Y361F, Y419F Mutant of Staphylococcus aureus CoADR | | Descriptor: | CHLORIDE ION, COENZYME A, Coenzyme A disulfide reductase, ... | | Authors: | Edwards, J.S, Wallace, B.D, Wallen, J.R, Claiborne, A, Redinbo, M.R. | | Deposit date: | 2012-04-19 | | Release date: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Turnover-Dependent Covalent Inactivation of Staphylococcus aureus Coenzyme A-Disulfide Reductase by Coenzyme A-Mimetics: Mechanistic and Structural Insights.
Biochemistry, 51, 2012
|
|
4EM4
 
 | | Crystal Structure of Staphylococcus aureus bound with the covalent inhibitor Pethyl-VS-CoA | | Descriptor: | CHLORIDE ION, Coenzyme A disulfide reductase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Wallace, B.D, Edwards, J.S, Claiborne, A, Redinbo, M.R. | | Deposit date: | 2012-04-11 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Turnover-Dependent Covalent Inactivation of Staphylococcus aureus Coenzyme A-Disulfide Reductase by Coenzyme A-Mimetics: Mechanistic and Structural Insights.
Biochemistry, 51, 2012
|
|
4EM3
 
 | | Crystal Structure of Staphylococcus aureus bound with the covalent inhibitor MeVS-CoA | | Descriptor: | CHLORIDE ION, Coenzyme A disulfide reductase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Wallace, B.D, Edwards, J.S, Claiborne, A, Redinbo, M.R. | | Deposit date: | 2012-04-11 | | Release date: | 2012-10-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Turnover-Dependent Covalent Inactivation of Staphylococcus aureus Coenzyme A-Disulfide Reductase by Coenzyme A-Mimetics: Mechanistic and Structural Insights.
Biochemistry, 51, 2012
|
|
4EQS
 
 | | Crystal structure of the Y419F mutant of Staphylococcus aureus CoADR | | Descriptor: | CHLORIDE ION, COENZYME A, Coenzyme A disulfide reductase, ... | | Authors: | Wallace, B.D, Edwards, J.S, Wallen, J.R, Claiborne, A, Redinbo, M.R. | | Deposit date: | 2012-04-19 | | Release date: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Turnover-Dependent Covalent Inactivation of Staphylococcus aureus Coenzyme A-Disulfide Reductase by Coenzyme A-Mimetics: Mechanistic and Structural Insights.
Biochemistry, 51, 2012
|
|
6MVH
 
 | |
6MVF
 
 | |
4J5X
 
 | | Crystal Structure of the SR12813-bound PXR/RXRalpha LBD Heterotetramer Complex | | Descriptor: | Nuclear receptor subfamily 1 group I member 2, Nuclear receptor coactivator 1, Retinoic acid receptor RXR-alpha, ... | | Authors: | Wallace, B.D, Betts, L, Redinbo, M.R. | | Deposit date: | 2013-02-10 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Functional Analysis of the Human Nuclear Xenobiotic Receptor PXR in Complex with RXRalpha.
J.Mol.Biol., 425, 2013
|
|
6MVG
 
 | |
6NCW
 
 | |
6NCX
 
 | |
6NZG
 
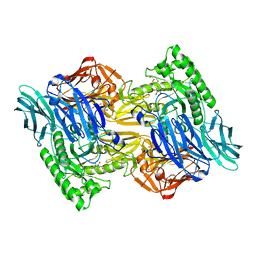 | | Bacteroides uniformis beta-glucuronidase 2 covalently bound to cyclophellitol-6-carboxylate aziridine | | Descriptor: | (1S,2R,3S,4S,5S,6R)-2-amino-3,4,5,6-tetrahydroxycyclohexane-1-carboxylic acid, Beta-galactosidase, CALCIUM ION, ... | | Authors: | Pellock, S.J, Jariwala, P.B, Redinbo, M.R. | | Deposit date: | 2019-02-13 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovering the Microbial Enzymes Driving Drug Toxicity with Activity-Based Protein Profiling.
Acs Chem.Biol., 15, 2020
|
|
4EQR
 
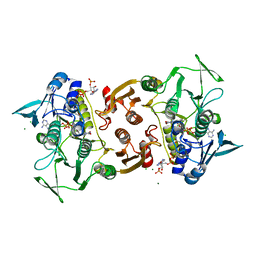 | | Crystal structure of the Y361F mutant of Staphylococcus aureus CoADR | | Descriptor: | CHLORIDE ION, COENZYME A, Coenzyme A disulfide reductase, ... | | Authors: | Wallace, B.D, Edwards, J.S, Wallen, J.R, Claiborne, A, Redinbo, M.R. | | Deposit date: | 2012-04-19 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Turnover-Dependent Covalent Inactivation of Staphylococcus aureus Coenzyme A-Disulfide Reductase by Coenzyme A-Mimetics: Mechanistic and Structural Insights.
Biochemistry, 51, 2012
|
|
4EMW
 
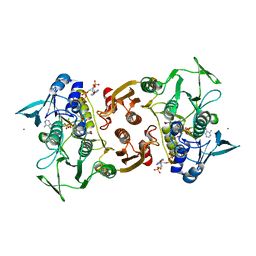 | | Crystal Structure of Staphylococcus aureus bound with the covalent inhibitor EtVC-CoA | | Descriptor: | CHLORIDE ION, Coenzyme A disulfide reductase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Edwards, J.S, Wallace, B.D, Claiborne, A, Redinbo, M.R. | | Deposit date: | 2012-04-12 | | Release date: | 2012-10-17 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Turnover-Dependent Covalent Inactivation of Staphylococcus aureus Coenzyme A-Disulfide Reductase by Coenzyme A-Mimetics: Mechanistic and Structural Insights.
Biochemistry, 51, 2012
|
|
4JKM
 
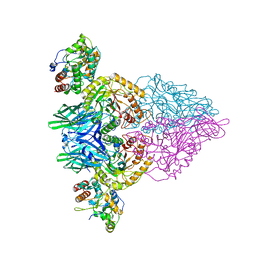 | |
4JKK
 
 | |
6NCZ
 
 | |
6NCY
 
 | | Crystal structure of hybrid beta-glucuronidase/beta-galacturonidase from Fusicatenibacter saccharivorans | | Descriptor: | Beta-glucuronidase, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2018-12-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Selecting a Single Stereocenter: The Molecular Nuances That Differentiate beta-Hexuronidases in the Human Gut Microbiome.
Biochemistry, 58, 2019
|
|
