5ZMC
 
 | | Structural Basis for Reactivation of -146C>T Mutant TERT Promoter by cooperative binding of p52 and ETS1/2 | | Descriptor: | DNA (5'-D(P*CP*GP*GP*GP*GP*AP*CP*CP*CP*GP*GP*AP*AP*GP*GP*G)-3'), DNA (5'-D(P*GP*CP*CP*CP*TP*TP*CP*CP*GP*GP*GP*TP*CP*CP*CP*C)-3'), Nuclear factor NF-kappa-B p100 subunit, ... | | Authors: | Xu, X, Bharath, S.R, Song, H. | | Deposit date: | 2018-04-02 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis for reactivating the mutant TERT promoter by cooperative binding of p52 and ETS1.
Nat Commun, 9, 2018
|
|
5FHG
 
 | |
5FHD
 
 | | Structure of Bacteroides sp Pif1 complexed with tailed dsDNA resulting in ssDNA bound complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*CP*CP*GP*GP*GP*GP*CP*CP*GP*CP*GP*C)-3'), MAGNESIUM ION, ... | | Authors: | Zhou, X, Ren, W, Bharath, S.R, Song, H. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Insights into the Unwinding Mechanism of Bacteroides sp Pif1
Cell Rep, 14, 2016
|
|
4EIH
 
 | | Crystal structure of Arg SH2 domain | | Descriptor: | Abelson tyrosine-protein kinase 2, CHLORIDE ION | | Authors: | Liu, W, MacGrath, S.M, Koleske, A.J, Boggon, T.J. | | Deposit date: | 2012-04-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Two Amino Acid Residues Confer Different Binding Affinities of Abelson Family Kinase Src Homology 2 Domains for Phosphorylated Cortactin.
J.Biol.Chem., 289, 2014
|
|
6YMY
 
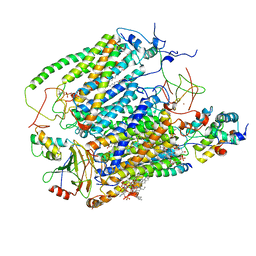 | | Cytochrome c oxidase from Saccharomyces cerevisiae | | Descriptor: | (2R,5S,11R,14R)-5,8,11-trihydroxy-2-(nonanoyloxy)-5,11-dioxido-16-oxo-14-[(propanoyloxy)methyl]-4,6,10,12,15-pentaoxa-5,11-diphosphanonadec-1-yl undecanoate, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, COPPER (II) ION, ... | | Authors: | Berndtsson, J, Rathore, S, Ott, M. | | Deposit date: | 2020-04-10 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Respiratory supercomplexes enhance electron transport by decreasing cytochrome c diffusion distance.
Embo Rep., 21, 2020
|
|
3USU
 
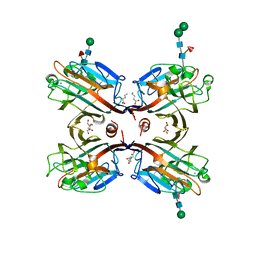 | | Crystal structure of Butea monosperma seed lectin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Abhilash, J, Geethanandan, K, Bharath, S.R, Sadasivan, C, Haridas, M. | | Deposit date: | 2011-11-24 | | Release date: | 2012-01-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal structure of Butea monosperma seed lectin
To be Published
|
|
7UN5
 
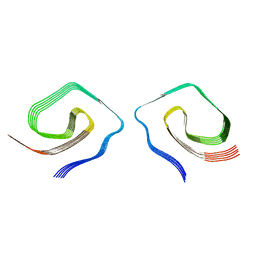 | |
7UMQ
 
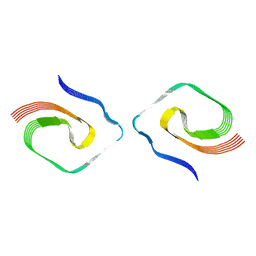 | |
7TMC
 
 | |
8SEL
 
 | |
3ULR
 
 | | Lysozyme contamination facilitates crystallization of a hetero-trimericCortactin:Arg:Lysozyme complex | | Descriptor: | Abelson tyrosine-protein kinase 2, Lysozyme C, Src substrate cortactin | | Authors: | Liu, W, MacGrath, S, Koleske, A.J, Boggon, T.J. | | Deposit date: | 2011-11-11 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Lysozyme contamination facilitates crystallization of a heterotrimeric cortactin-Arg-lysozyme complex.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
6YMX
 
 | | CIII2/CIV respiratory supercomplex from Saccharomyces cerevisiae | | Descriptor: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (1R)-2-(phosphonooxy)-1-[(tridecanoyloxy)methyl]ethyl pentadecanoate, (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(heptanoyloxy)methyl]ethyl octadecanoate, ... | | Authors: | Berndtsson, J, Rathore, S, Ott, M. | | Deposit date: | 2020-04-10 | | Release date: | 2020-09-09 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Respiratory supercomplexes enhance electron transport by decreasing cytochrome c diffusion distance.
Embo Rep., 21, 2020
|
|
7X0E
 
 | | Structure of Pseudomonas NRPS protein, AmbB-TC in apo form | | Descriptor: | AMB antimetabolite synthase AmbB, N-methyl-N-[(2S,3R,4R,5R)-2,3,4,5,6-pentakis(oxidanyl)hexyl]nonanamide | | Authors: | ChuYuanKee, M, Bharath, S.R, Song, H. | | Deposit date: | 2022-02-22 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the substrate-bound condensation domains of non-ribosomal peptide synthetase AmbB.
Sci Rep, 12, 2022
|
|
6L3G
 
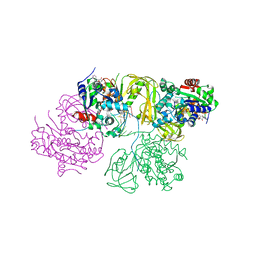 | | Structural Basis for DNA Unwinding at Forked dsDNA by two coordinating Pif1 helicases | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*TP*TP*TP*T)-3'), ... | | Authors: | Su, N, Bharath, S.R, Song, H. | | Deposit date: | 2019-10-10 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for DNA unwinding at forked dsDNA by two coordinating Pif1 helicases.
Nat Commun, 10, 2019
|
|
9CZI
 
 | |
9CZL
 
 | |
3OZZ
 
 | |
7JG1
 
 | | Dimeric Immunoglobin A (dIgA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Igh protein, ... | | Authors: | Kumar Bharathkar, S, Parker, B.P, Malyutin, A.G, Stadtmueller, B.M. | | Deposit date: | 2020-07-18 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structures of Secretory and dimeric Immunoglobulin A.
Elife, 9, 2020
|
|
7JG2
 
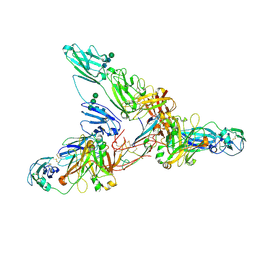 | | Secretory Immunoglobin A (SIgA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Igh protein, ... | | Authors: | Kumar Bharathkar, S, Parker, B.P, Malyutin, A.G, Stadtmueller, B.M. | | Deposit date: | 2020-07-18 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structures of Secretory and dimeric Immunoglobulin A.
Elife, 9, 2020
|
|
7X0F
 
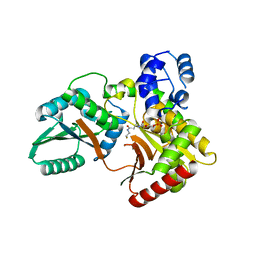 | |
6LG2
 
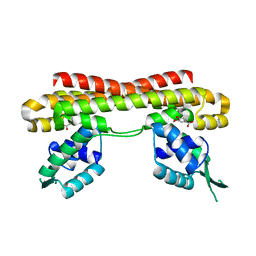 | | VanR bound to Vanillate | | Descriptor: | 4-HYDROXY-3-METHOXYBENZOATE, Predicted transcriptional regulators | | Authors: | He, Y, Bharath, S.R, Song, H. | | Deposit date: | 2019-12-04 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Developing a highly efficient hydroxytyrosol whole-cell catalyst by de-bottlenecking rate-limiting steps.
Nat Commun, 11, 2020
|
|
9CZN
 
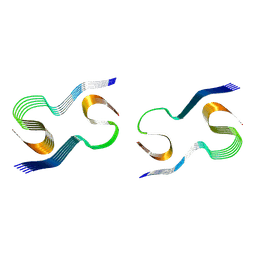 | |
9CZP
 
 | |
2ZLF
 
 | | The Structural Basis for Peptidomimetic Inhibition of Eukaryotic Ribonucleotide Reductase | | Descriptor: | FTLDADF, Ribonucleoside-diphosphate reductase large chain 1 | | Authors: | Xu, H, Fairman, J.W, Wijerathna, S.R, LaMacchia, J, Kreischer, N.R, Helmbrecht, E, Cooperman, B.S, Dealwis, C. | | Deposit date: | 2008-04-09 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | The Structural Basis for Peptidomimetic Inhibition of Eukaryotic Ribonucleotide Reductase: A Conformationally Flexible Pharmacophore
J.Med.Chem., 51, 2008
|
|
4JF0
 
 | | N79R mutant of N-acetylornithine aminotransferase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Acetylornithine/succinyldiaminopimelate aminotransferase, ... | | Authors: | Bisht, S, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational transitions, ligand specificity and catalysis in N-acetylornithine aminotransferase: Implications on drug designing and rational enzyme engineering in omega aminotransferases
To be Published
|
|
