7SYE
 
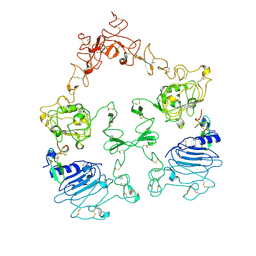 | | Cryo-EM structure of the extracellular module of the full-length EGFR bound to EGF. "tips-separated" conformation | | Descriptor: | Epidermal growth factor, Epidermal growth factor receptor | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7SZ0
 
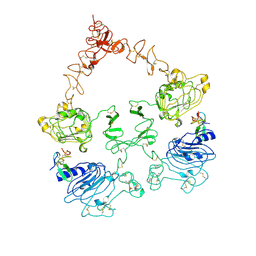 | | Cryo-EM structure of the extracellular module of the full-length EGFR L834R bound to EGF. "tips-juxtaposed" conformation | | Descriptor: | Epidermal growth factor, Epidermal growth factor receptor | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-25 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7UY4
 
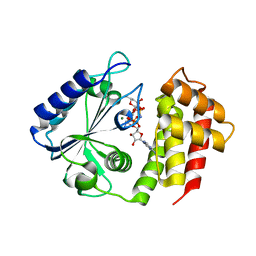 | | Aminoglycoside-modifying enzyme ANT-3,9 in complex with spectinomycin and AMP-PNP | | Descriptor: | Aminoglycoside (3'') (9) adenylyltransferase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Reeve, S.M, Lee, R.E, Das, N, Jayaraman, S. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Characterization of the spectinomycin deactivating enzyme ANT-3,9
To Be Published
|
|
6BUZ
 
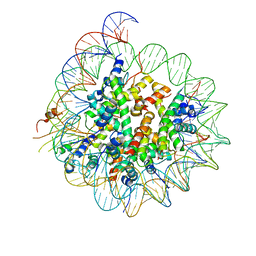 | | Cryo-EM structure of CENP-A nucleosome in complex with kinetochore protein CENP-N | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B, ... | | Authors: | Chittori, S, Hong, J, Kelly, A.E, Bai, Y, Subramaniam, S. | | Deposit date: | 2017-12-11 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP-N.
Science, 359, 2018
|
|
7LAD
 
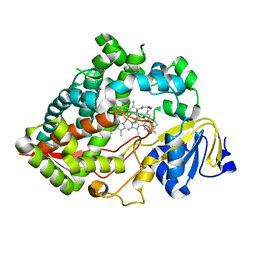 | | Clobetasol propionate bound to CYP3A5 | | Descriptor: | Clobetasol propionate, Cytochrome P450 3A5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Buchman, C.D, Miller, D, Wang, J, Jayaraman, S, Chen, T. | | Deposit date: | 2021-01-06 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Unraveling the Structural Basis of Selective Inhibition of Human Cytochrome P450 3A5.
J.Am.Chem.Soc., 143, 2021
|
|
2VDA
 
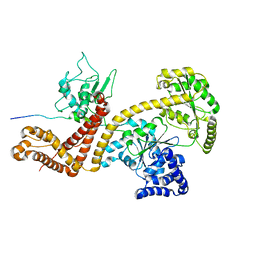 | | Solution structure of the SecA-signal peptide complex | | Descriptor: | MALTOPORIN, TRANSLOCASE SUBUNIT SECA | | Authors: | Gelis, I, Bonvin, A.M.J.J, Keramisanou, D, Koukaki, M, Gouridis, G, Karamanou, S, Economou, A, Kalodimos, C.G. | | Deposit date: | 2007-10-01 | | Release date: | 2007-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Signal-Sequence Recognition by the Translocase Motor Seca as Determined by NMR
Cell(Cambridge,Mass.), 131, 2007
|
|
6AC7
 
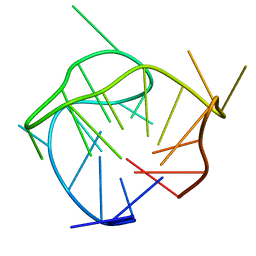 | | Structure of a (3+1) hybrid G-quadruplex in the PARP1 promoter | | Descriptor: | 5'-D(*TP*GP*GP*GP*GP*TP*CP*CP*GP*AP*GP*GP*CP*GP*GP*GP*GP*CP*TP*TP*GP*GP*G)-3' | | Authors: | Sengar, A, Vandana, J.J, Chambers, V.S, Di Antonio, M, Winnerdy, F.R, Balasubramanian, S, Phan, A.T. | | Deposit date: | 2018-07-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a (3+1) hybrid G-quadruplex in the PARP1 promoter.
Nucleic Acids Res., 47, 2019
|
|
6B44
 
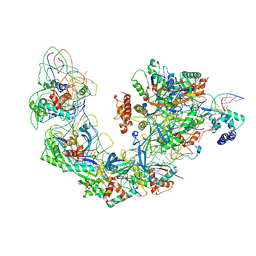 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound target dsDNA | | Descriptor: | CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, CRISPR-associated protein Csy2, ... | | Authors: | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | Deposit date: | 2017-09-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6B45
 
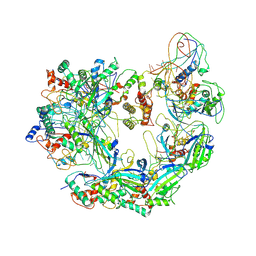 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex | | Descriptor: | CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, CRISPR-associated protein Csy2, ... | | Authors: | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | Deposit date: | 2017-09-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6B48
 
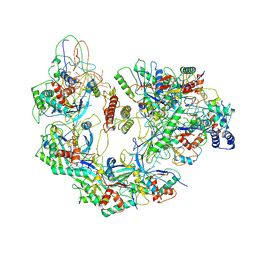 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound anti-CRISPR protein AcrF10 | | Descriptor: | Anti-CRISPR protein AcrF10, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | Deposit date: | 2017-09-25 | | Release date: | 2017-10-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6B47
 
 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound anti-CRISPR protein AcrF2 | | Descriptor: | Anti-CRISPR protein AcrF2, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | Deposit date: | 2017-09-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
4CC8
 
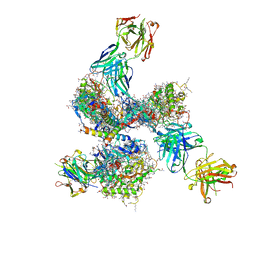 | | Pre-fusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy | | Descriptor: | GP120, GP41, MONOCLONAL ANTIBODY VRC03 FAB HEAVY CHAIN, ... | | Authors: | Bartesaghi, A, Merk, A, Borgnia, M.J, Milne, J.L.S, Subramaniam, S. | | Deposit date: | 2013-10-18 | | Release date: | 2013-10-30 | | Last modified: | 2018-01-10 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Prefusion Structure of Trimeric HIV-1 Envelope Glycoprotein Determined by Cryo-Electron Microscopy.
Nat.Struct.Mol.Biol., 20, 2013
|
|
1ZB1
 
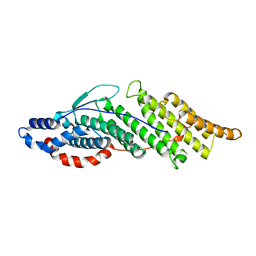 | | Structure basis for endosomal targeting by the Bro1 domain | | Descriptor: | BRO1 protein | | Authors: | Kim, J, Sitaraman, S, Hierro, A, Beach, B.M, Odorizzi, G, Hurley, J.H. | | Deposit date: | 2005-04-07 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for endosomal targeting by the Bro1 domain.
Dev.Cell, 8, 2005
|
|
2KQH
 
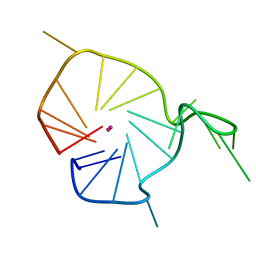 | | A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics | | Descriptor: | 5'-D(*CP*GP*GP*GP*CP*GP*GP*GP*CP*GP*CP*GP*AP*GP*GP*GP*AP*GP*GP*GP*T)-3', POTASSIUM ION | | Authors: | Hsu, S.-T.D, Varnai, P, Bugaut, A, Reszka, A.P, Neidle, S, Balasubramanian, S. | | Deposit date: | 2009-11-05 | | Release date: | 2009-11-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics
J.Am.Chem.Soc., 131, 2009
|
|
6B46
 
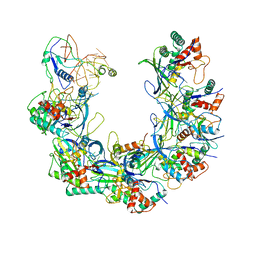 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound anti-CRISPR protein AcrF1 | | Descriptor: | Anti-CRISPR protein AcrF1, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy3, ... | | Authors: | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | Deposit date: | 2017-09-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
2YOJ
 
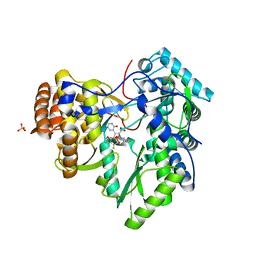 | | HCV NS5B polymerase complexed with pyridonylindole compound | | Descriptor: | 4-fluoranyl-6-[(7-fluoranyl-4-oxidanylidene-3H-quinazolin-6-yl)methyl]-8-(2-oxidanylidene-1H-pyridin-3-yl)furo[2,3-e]indole-7-carboxylic acid, PHOSPHATE ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Chen, K.X, Venkatraman, S, Anilkumar, G.N, Zeng, Q, Lesburg, C.A, Vibulbhan, B, Yang, W, Velazquez, F, Chan, T.-Y, Bennett, F, Sannigrahi, M, Jiang, Y, Duca, J.S, Pinto, P, Gavalas, S, Huang, Y, Wu, W, Selyutin, O, Agrawal, S, Feld, B, Huang, H.-C, Li, C, Cheng, K.-C, Shih, N.-Y, Kozlowski, J.A, Rosenblum, S.B, Njoroge, F.G. | | Deposit date: | 2012-10-24 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery of Sch 900188: A Potent Hepatitis C Virus Ns5B Polymerase Inhibitor Prodrug as a Development Candidate
Acs Med.Chem.Lett., 5, 2014
|
|
2KQG
 
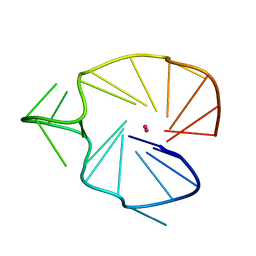 | | A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics | | Descriptor: | 5'-D(*CP*GP*GP*GP*CP*GP*GP*GP*CP*AP*CP*GP*AP*GP*GP*GP*AP*GP*GP*GP*T)-3', POTASSIUM ION | | Authors: | Hsu, S.-T.D, Varnai, P, Bugaut, A, Reszka, A.P, Neidle, S, Balasubramanian, S. | | Deposit date: | 2009-11-05 | | Release date: | 2009-11-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics
J.Am.Chem.Soc., 131, 2009
|
|
3MXB
 
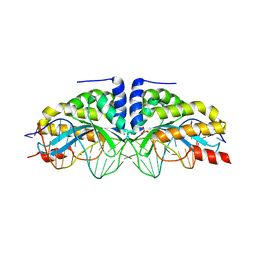 | | Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*CP*TP*GP*GP*CP*TP*GP*AP*GP*GP*TP*AP*CP*CP*TP*GP*AP*GP*AP*AP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*TP*TP*CP*TP*CP*AP*GP*GP*TP*AP*CP*CP*TP*CP*AP*GP*CP*CP*AP*GP*A)-3'), ... | | Authors: | Munoz, I.G, Prieto, J, Subramanian, S, Coloma, J, Montoya, G. | | Deposit date: | 2010-05-07 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus.
Nucleic Acids Res., 39, 2011
|
|
3U4R
 
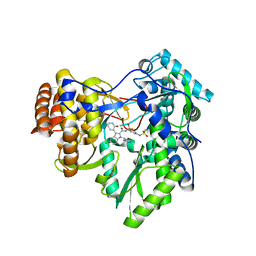 | | Novel HCV NS5B polymerase Inhibitors: Discovery of Indole C2 Acyl sulfonamides | | Descriptor: | 1-[(2-aminopyridin-4-yl)methyl]-5-chloro-N-({3-[(methylsulfonyl)amino]phenyl}sulfonyl)-3-(2-oxo-1,2-dihydropyridin-3-yl)-1H-indole-2-carboxamide, RNA-directed RNA polymerase | | Authors: | Anilkumar, G.N, Selyutin, O, Rosenblum, S.B, Zeng, Q, Jiang, Y, Chan, T.-Y, Pu, H, Wang, L, Bennett, F, Chen, K.X, Lesburg, C.A, Duca, J.S, Gavalas, S, Huang, Y, Pinto, P, Sannagrahi, M, Velazquez, F, Venkataraman, S, Vilbubhan, B, Agrawal, S, Ferrari, E, Jiang, C.-K, Huang, H.-C, Shih, N.-Y, Njoroge, F.G, Kozlowski, J.A. | | Deposit date: | 2011-10-10 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | II. Novel HCV NS5B polymerase inhibitors: Discovery of indole C2 acyl sulfonamides.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3U4O
 
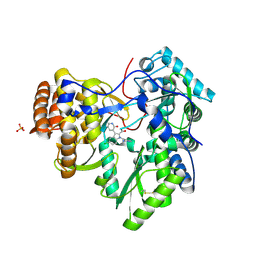 | | Novel HCV NS5B polymerase Inhibitors: Discovery of Indole C2 Acyl sulfonamides | | Descriptor: | 1-[(2-aminopyridin-4-yl)methyl]-5-chloro-3-(2-oxo-1,2-dihydropyridin-3-yl)-1H-indole-2-carboxylic acid, PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Anilkumar, G.N, Selyutin, O, Rosenblum, S.B, Zeng, Q, Jiang, Y, Chan, T.-Y, Pu, H, Wang, L, Bennett, F, Chen, K.X, Lesburg, C.A, Duca, J.S, Gavalas, S, Huang, Y, Pinto, P, Sannigrahi, M, Velazquez, F, Venkataraman, S, Vilbubhan, B, Agrawal, S, Ferrari, E, Jiang, C.-K, Huang, H.-C, Shih, N.-Y, Njoroge, F.G, Kozlowski, J.A. | | Deposit date: | 2011-10-10 | | Release date: | 2011-12-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | II. Novel HCV NS5B polymerase inhibitors: Discovery of indole C2 acyl sulfonamides.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1YX9
 
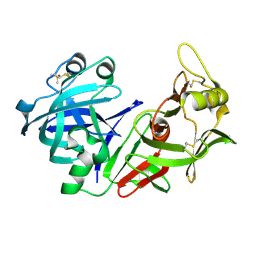 | |
3N90
 
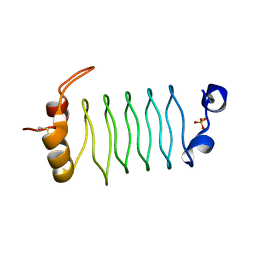 | | The 1.7 Angstrom resolution crystal structure of AT2G44920, a pentapeptide repeat protein from Arabidopsis thaliana thylakoid lumen. | | Descriptor: | SULFATE ION, Thylakoid lumenal 15 kDa protein 1, chloroplastic | | Authors: | Ni, S, Mckgookey, M, Tinch, S.L, Jones, A.N, Jayaraman, S, Kennedy, M.A. | | Deposit date: | 2010-05-28 | | Release date: | 2011-06-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7 Angstrom resolution crystal structure of AT2G44920, a pentapeptide repeat protein from Arabidopsis thaliana thylakoid lumen.
To be Published
|
|
3UPI
 
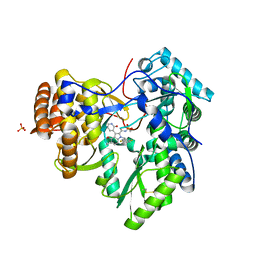 | | Synthesis of novel 4,5-dihydrofurano indoles and their evaluation as HCV NS5B polymerase inhibitors | | Descriptor: | (3S)-6-(2,5-difluorobenzyl)-3-methyl-N-(methylsulfonyl)-8-(2-oxo-1,2-dihydropyridin-3-yl)-3,6-dihydro-2H-furo[2,3-e]indole-7-carboxamide, PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Velazquez, F, Venkataraman, S, Lesburg, C.A, Duca, J.S, Rosenblum, S.B, Kozlowski, J.A, Njoroge, F.G. | | Deposit date: | 2011-11-18 | | Release date: | 2012-01-25 | | Last modified: | 2012-02-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis of New 4,5-Dihydrofuranoindoles and Their Evaluation as HCV NS5B Polymerase Inhibitors.
Org.Lett., 14, 2012
|
|
3UPH
 
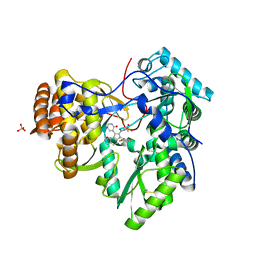 | | Synthesis of novel 4,5-dihydrofurano indoles and their evaluation as HCV NS5B polymerase inhibitors | | Descriptor: | 6-(2,5-difluorobenzyl)-N-(methylsulfonyl)-8-(2-oxo-1,2-dihydropyridin-3-yl)-3,6-dihydro-2H-furo[2,3-e]indole-7-carboxamide, PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Velazquez, F, Venkataraman, S, Lesburg, C.A, Duca, J.S, Rosenblum, S.B, Kozlowski, J.A, Njoroge, F.G. | | Deposit date: | 2011-11-18 | | Release date: | 2012-01-25 | | Last modified: | 2012-02-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis of New 4,5-Dihydrofuranoindoles and Their Evaluation as HCV NS5B Polymerase Inhibitors.
Org.Lett., 14, 2012
|
|
2G8T
 
 | |
