3GTV
 
 | |
1Z0H
 
 | | N-terminal helix reorients in recombinant C-fragment of Clostridium botulinum type B | | 分子名称: | Botulinum neurotoxin type B | | 著者 | Jayaraman, S, Eswarmoorthy, S, Ashraf, S.A, Smith, L.A, Swaminathan, S. | | 登録日 | 2005-03-01 | | 公開日 | 2005-03-15 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | N-terminal helix reorients in recombinant C-fragment of Clostridium botulinum type B.
Biochem.Biophys.Res.Commun., 330, 2005
|
|
3KNX
 
 | | HCV NS3 protease domain with P1-P3 macrocyclic ketoamide inhibitor | | 分子名称: | (2R)-2-{(3S,13S,16aS,17aR,17bS)-13-[({(1S)-1-[(4,4-dimethyl-2,6-dioxopiperidin-1-yl)methyl]-2,2-dimethylpropyl}carbamoyl)amino]-17,17-dimethyl-1,14-dioxooctadecahydro-2H-cyclopropa[3,4]pyrrolo[1,2-a][1,4]diazacyclohexadecin-3-yl}-2-hydroxy-N-prop-2-en-1-ylethanamide, BETA-MERCAPTOETHANOL, HCV NS3 Protease, ... | | 著者 | Venkatraman, S, Velazquez, F, Wu, W, Blackman, M, Chen, K.X, Bogen, S, Nair, L, Tong, X, Chase, R, Hart, A, Agrawal, S, Pichardo, J, Prongay, A, Cheng, K.-C, Girijavallabhan, V, Piwinski, J, Shih, N.-Y, Njoroge, F.G. | | 登録日 | 2009-11-12 | | 公開日 | 2010-10-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Discovery and structure-activity relationship of P1-P3 ketoamide derived macrocyclic inhibitors of hepatitis C virus NS3 protease.
J.Med.Chem., 52, 2009
|
|
3H2P
 
 | | Human SOD1 D124V Variant | | 分子名称: | ACETYL GROUP, MALONATE ION, Superoxide dismutase [Cu-Zn], ... | | 著者 | Seetharaman, S.V, Winkler, D.D, Taylor, A.B, Cao, X, Whitson, L.J, Doucette, P.A, Valentine, J.S, Carroll, M.C, Culotta, V.C, Hart, P.J. | | 登録日 | 2009-04-14 | | 公開日 | 2010-05-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structures of Pathogenic SOD1 Mutants H80R and D124V: Disrupted Zinc-binding
and Compromised Post-translational Modification by the Copper Chaperone CCS
To be Published
|
|
3GTT
 
 | | Mouse SOD1 | | 分子名称: | Superoxide dismutase [Cu-Zn], ZINC ION | | 著者 | Seetharaman, S.V, Taylor, A.B, Hart, P.J. | | 登録日 | 2009-03-28 | | 公開日 | 2010-09-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structures of mouse SOD1 and human/mouse SOD1 chimeras.
Arch.Biochem.Biophys., 503, 2010
|
|
3H2Q
 
 | | Human SOD1 H80R variant, P21 crystal form | | 分子名称: | SULFATE ION, Superoxide dismutase [Cu-Zn], ZINC ION | | 著者 | Seetharaman, S.V, Winkler, D.D, Taylor, A.B, Cao, X, Whitson, L.J, Doucette, P.A, Valentine, J.S, Carroll, M.C, Culotta, V.C, Hart, P.J. | | 登録日 | 2009-04-14 | | 公開日 | 2010-05-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structures of Pathogenic SOD1 Mutants H80R and D124V: Disrupted Zinc-binding
and Compromised Post-translational Modification by the Copper Chaperone CCS
To be Published
|
|
1YXW
 
 | | A common binding site for disialyllactose and a tri-peptide in the C-fragment of tetanus neurotoxin | | 分子名称: | GLUTAMIC ACID, TRYPTOPHAN, TYROSINE, ... | | 著者 | Jayaraman, S, Eswaramoorthy, S, Kumaran, D, Swaminathan, S. | | 登録日 | 2005-02-22 | | 公開日 | 2005-03-15 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Common binding site for disialyllactose and tri-peptide in C-fragment of tetanus neurotoxin
Proteins, 61, 2005
|
|
3QQD
 
 | | Human SOD1 H80R variant, P212121 crystal form | | 分子名称: | SULFATE ION, Superoxide dismutase [Cu-Zn], ZINC ION | | 著者 | Seetharaman, S.V, Winkler, D.D, Taylor, A.B, Cao, X, Whitson, L.J, Doucette, P.A, Valentine, J.S, Schirf, V, Demeler, B, Carroll, M.C, Culotta, V.C, Hart, P.J. | | 登録日 | 2011-02-15 | | 公開日 | 2011-03-09 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.653 Å) | | 主引用文献 | Disrupted zinc-binding sites in structures of pathogenic SOD1 variants D124V and H80R.
Biochemistry, 49, 2010
|
|
2F9U
 
 | | HCV NS3 protease domain with NS4a peptide and a ketoamide inhibitor with a P2 norborane | | 分子名称: | 1,1-DIMETHYLETHYL [1-CYCLOHEXYL-2-[3-[[[1-[2-[[2-[[2-(DIMETHYLAMINO)-2-OXO-1-PHENYLETHYL]AMINO]-2-OXOETHYL]AMINO]-1,2-DIOXOETHYL]PENTYL]AMINO]CARBONYL]-2-AZABICYCLO[2.2.1]HEPTAN-2-YL]-2-OXOETHYL]CARBAMATE, NS3 protease/helicase', ZINC ION, ... | | 著者 | Venkatraman, S, Njoroge, F.G, Wu, W, Girijavallabhan, V, Prongay, A.J, Butkiewicz, N, Pichardo, J. | | 登録日 | 2005-12-06 | | 公開日 | 2006-06-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Novel Inhibitors of Hepatitis C NS3-NS4A Serine Protease Derived from 2-Aza-bicyclo[2.2.1]heptane-3-carboxylic acid.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1FBK
 
 | |
1FBB
 
 | |
3EYD
 
 | | Structure of HCV NS3-4A Protease with an Inhibitor Derived from a Boronic Acid | | 分子名称: | HCV NS3, HCV NS4a peptide, ZINC ION, ... | | 著者 | Venkatraman, S, Wu, W, Prongay, A.J, Girijavallabhan, V, Njoroge, F.G. | | 登録日 | 2008-10-20 | | 公開日 | 2009-02-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Potent inhibitors of HCV-NS3 protease derived from boronic acids.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
8ILU
 
 | | Crystal structure of mouse Galectin-3 in complex with small molecule inhibitor | | 分子名称: | (2R,3R,4R,5R,6S)-2-(hydroxymethyl)-6-[2-(2-methyl-1,3-benzothiazol-6-yl)-1,2,4-triazol-3-yl]-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3, SODIUM ION, ... | | 著者 | Kumar, A, Jinal, S, Raman, S, Ghosh, K. | | 登録日 | 2023-03-04 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-09-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Identification of benzothiazole derived monosaccharides as potent, selective, and orally bioavailable inhibitors of human and mouse galectin-3; a rare example of using a S···O binding interaction for drug design.
Bioorg.Med.Chem., 101, 2024
|
|
7KD8
 
 | | TtgR C137I I141W M167L F168Y mutant in complex with resveratrol | | 分子名称: | HTH-type transcriptional regulator TtgR, MAGNESIUM ION, RESVERATROL | | 著者 | Bingman, C.A, Nishikawa, K.K, Smith, R.W, Raman, S. | | 登録日 | 2020-10-08 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Epistasis shapes the fitness landscape of an allosteric specificity switch.
Nat Commun, 12, 2021
|
|
7K1A
 
 | | TtgR quadruple mutant (C137I I141W M167L F168Y) | | 分子名称: | HTH-type transcriptional regulator TtgR, MAGNESIUM ION | | 著者 | Bingman, C.A, Nishikawa, K.K, Smith, R.W, Raman, S. | | 登録日 | 2020-09-07 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Epistasis shapes the fitness landscape of an allosteric specificity switch.
Nat Commun, 12, 2021
|
|
7K1C
 
 | | TtgR in complex with resveratrol | | 分子名称: | HTH-type transcriptional regulator TtgR, MAGNESIUM ION, RESVERATROL | | 著者 | Bingman, C.A, Nishikawa, K.K, Smith, R.W, Raman, S. | | 登録日 | 2020-09-07 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Epistasis shapes the fitness landscape of an allosteric specificity switch.
Nat Commun, 12, 2021
|
|
2L15
 
 | | Solution Structure of Cold Shock Protein CspA Using Combined NMR and CS-Rosetta method | | 分子名称: | Cold shock protein CspA | | 著者 | Tang, Y, Schneider, W.M, Shen, Y, Raman, S, Inouye, M, Baker, D, Roth, M.J, Montelione, G.T. | | 登録日 | 2010-07-22 | | 公開日 | 2010-09-15 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Fully automated high-quality NMR structure determination of small (2)H-enriched proteins.
J Struct Funct Genomics, 11, 2010
|
|
7XFA
 
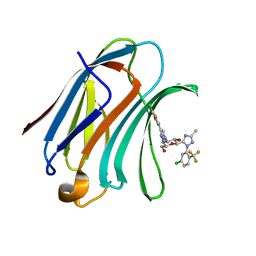 | | Structure of human Galectin-3 CRD in complex with monosaccharide inhibitor | | 分子名称: | (2~{S},3~{R},4~{R},5~{R},6~{R})-4-[4-[4-chloranyl-3,5-bis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]-2-[2-[5-chloranyl-2-(trifluoromethyl)phenyl]-5-methyl-1,2,4-triazol-3-yl]-6-(hydroxymethyl)oxane-3,5-diol, Galectin-3 | | 著者 | Shukla, J, Raman, S, Ghosh, K. | | 登録日 | 2022-04-01 | | 公開日 | 2022-10-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Identification of Monosaccharide Derivatives as Potent, Selective, and Orally Bioavailable Inhibitors of Human and Mouse Galectin-3.
J.Med.Chem., 65, 2022
|
|
6VYF
 
 | | Cryo-EM structure of Plasmodium vivax hexokinase (Open state) | | 分子名称: | Phosphotransferase | | 著者 | Srivastava, S.S, Darling, J.E, Suryadi, J, Morris, J.C, Drew, M.E, Subramaniam, S. | | 登録日 | 2020-02-26 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Plasmodium vivax and human hexokinases share similar active sites but display distinct quaternary architectures
Iucrj, 7, 2020
|
|
6CMO
 
 | | Rhodopsin-Gi complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab Heavy chain, Fab light chain, ... | | 著者 | Kang, Y, Kuybeda, O, de Waal, P.W, Mukherjee, S, Van Eps, N, Dutka, P, Zhou, X.E, Bartesaghi, A, Erramilli, S, Morizumi, T, Gu, X, Yin, Y, Liu, P, Jiang, Y, Meng, X, Zhao, G, Melcher, K, Earnst, O.P, Kossiakoff, A.A, Subramaniam, S, Xu, H.E. | | 登録日 | 2018-03-05 | | 公開日 | 2018-06-20 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Cryo-EM structure of human rhodopsin bound to an inhibitory G protein.
Nature, 558, 2018
|
|
6CGV
 
 | | Revised crystal structure of human adenovirus | | 分子名称: | Hexon protein, Hexon-interlacing protein, Penton protein, ... | | 著者 | Natchiar, S.K, Venkataraman, S, Nemerow, G.R, Reddy, V.S. | | 登録日 | 2018-02-21 | | 公開日 | 2018-04-25 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Revised Crystal Structure of Human Adenovirus Reveals the Limits on Protein IX Quasi-Equivalence and on Analyzing Large Macromolecular Complexes.
J. Mol. Biol., 430, 2018
|
|
5A1A
 
 | | 2.2 A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor | | 分子名称: | 2-phenylethyl 1-thio-beta-D-galactopyranoside, BETA-GALACTOSIDASE, MAGNESIUM ION, ... | | 著者 | Bartesaghi, A, Merk, A, Banerjee, S, Matthies, D, Wu, X, Milne, J, Subramaniam, S. | | 登録日 | 2015-04-29 | | 公開日 | 2015-05-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with a Cell-Permeant Inhibitor
Science, 348, 2015
|
|
3CQP
 
 | | Human SOD1 G85R Variant, Structure I | | 分子名称: | COPPER (II) ION, MALONATE ION, Superoxide dismutase [Cu-Zn], ... | | 著者 | Cao, X, Antonyuk, S, Seetharaman, S.V, Whitson, L.J, Taylor, A.B, Holloway, S.P, Strange, R.W, Doucette, P.A, Valentine, J.S, Tiwari, A, Hayward, L.J, Padua, S, Cohlberg, J.A, Hasnain, S.S, Hart, P.J. | | 登録日 | 2008-04-03 | | 公開日 | 2008-04-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structures of the G85R Variant of SOD1 in Familial Amyotrophic Lateral Sclerosis.
J.Biol.Chem., 283, 2008
|
|
2BDQ
 
 | | Crystal Structure of the Putative Copper Homeostasis Protein CutC from Streptococcus agalactiae, Northeast Strucural Genomics Target SaR15. | | 分子名称: | copper homeostasis protein CutC | | 著者 | Forouhar, F, Abashidze, M, Jayaraman, S, Ho, C.K, Cooper, B, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2005-10-20 | | 公開日 | 2005-11-01 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structure of the Putative Copper Homeostasis Protein CutC from Streptococcus agalactiae, Northeast Strucural Genomics Target SaR15.
To be Published
|
|
7SY7
 
 | | Cryo-EM structure of the SARS-CoV-2 D614G,N501Y,E484K,K417T mutant spike protein ectodomain bound to human ACE2 ectodomain (global refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | 登録日 | 2021-11-24 | | 公開日 | 2021-12-29 | | 実験手法 | ELECTRON MICROSCOPY (2.81 Å) | | 主引用文献 | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
