3L0U
 
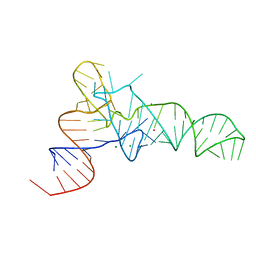 | | The crystal structure of unmodified tRNAPhe from Escherichia coli | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, Unmodified tRNAPhe | | Authors: | Byrne, R.T, Konevega, A.L, Rodnina, M.V, Antson, A.A. | | Deposit date: | 2009-12-10 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of unmodified tRNAPhe from Escherichia coli
Nucleic Acids Res., 38, 2010
|
|
4W4R
 
 | | Crystal structure of ent-kaurene synthase BJKS from bradyrhizobium japonicum | | Descriptor: | Uncharacterized protein blr2150 | | Authors: | Liu, W, Zheng, Y, Huang, C.H, Ko, T.P, Guo, R.T. | | Deposit date: | 2014-08-15 | | Release date: | 2015-01-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure, function and inhibition of ent-kaurene synthase from Bradyrhizobium japonicum.
Sci Rep, 4, 2014
|
|
3L0E
 
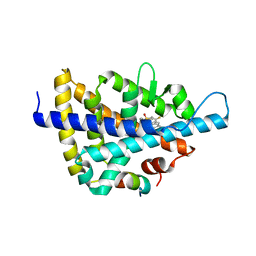 | | X-ray crystal structure of a Potent Liver X Receptor Modulator | | Descriptor: | N-(2-chloro-6-fluorobenzyl)-1-methyl-N-{[3'-(methylsulfonyl)biphenyl-4-yl]methyl}-1H-imidazole-4-sulfonamide, Nuclear receptor coactivator 2, Oxysterols receptor LXR-beta | | Authors: | Gampe Jr, R.T. | | Deposit date: | 2009-12-09 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of tertiary sulfonamides as potent liver X receptor antagonists.
J.Med.Chem., 53, 2010
|
|
2PXT
 
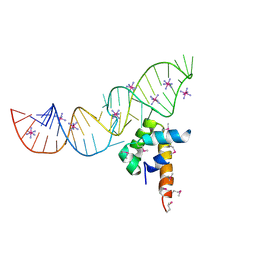 | | Variant 15 of Ribonucleoprotein Core of the E. Coli Signal Recognition Particle | | Descriptor: | 4.5 S RNA, COBALT HEXAMMINE(III), Signal recognition particle protein | | Authors: | Keel, A.Y, Rambo, R.P, Batey, R.T, Kieft, J.S. | | Deposit date: | 2007-05-14 | | Release date: | 2007-08-07 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A General Strategy to Solve the Phase Problem in RNA Crystallography.
Structure, 15, 2007
|
|
2PXF
 
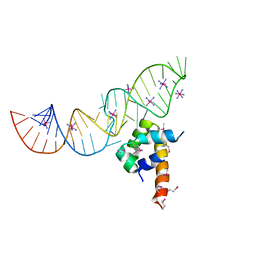 | | Variant 5 of Ribonucleoprotein Core of the E. Coli Signal Recognition Particle | | Descriptor: | 4.5 S RNA, COBALT HEXAMMINE(III), Signal recognition particle protein | | Authors: | Keel, A.Y, Rambo, R.P, Batey, R.T, Kieft, J.S. | | Deposit date: | 2007-05-14 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A General Strategy to Solve the Phase Problem in RNA Crystallography.
Structure, 15, 2007
|
|
2Q7K
 
 | |
2PXU
 
 | | Variant 16 of Ribonucleoprotein Core of the E. Coli Signal Recognition Particle | | Descriptor: | 4.5 S RNA, COBALT HEXAMMINE(III), Signal recognition particle protein | | Authors: | Keel, A.Y, Rambo, R.P, Batey, R.T, Kieft, J.S. | | Deposit date: | 2007-05-14 | | Release date: | 2007-08-07 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A General Strategy to Solve the Phase Problem in RNA Crystallography.
Structure, 15, 2007
|
|
2Q7L
 
 | |
4WOQ
 
 | | Crystal Structures of CdNal from Clostridium difficile in complex with ketobutyric | | Descriptor: | 2-KETOBUTYRIC ACID, N-acetylneuraminate lyase | | Authors: | Liu, W.D, Guo, R.T, Cui, Y.F, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2014-10-16 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of CdNal from Clostridium difficile in complex with ketobutyric
to be published
|
|
4WOZ
 
 | | Crystal Structures of CdNal from Clostridium difficile in complex with mannosamine | | Descriptor: | 2-(ACETYLAMINO)-2-DEOXY-D-MANNOSE, N-acetylneuraminate lyase | | Authors: | Liu, W.D, Guo, R.T, Cui, Y.F, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2014-10-17 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structures of CdNal from Clostridium difficile in complex with mannosamine
to be published
|
|
2Q7J
 
 | |
2QAZ
 
 | |
3LH1
 
 | | Q191A mutant of the DegS-deltaPDZ | | Descriptor: | Protease degS | | Authors: | Sohn, J, Grant, R.A, Sauer, R.T. | | Deposit date: | 2010-01-21 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | Allostery is an intrinsic property of the protease domain of DegS: implications for enzyme function and evolution.
J.Biol.Chem., 285, 2010
|
|
8E7V
 
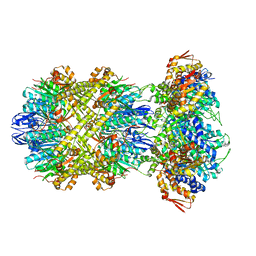 | | Cryo-EM structure of substrate-free DNClpX.ClpP from singly capped particles | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | Authors: | Ghanbarpour, A, Cohen, S, Davis, J.H, Sauer, R.T. | | Deposit date: | 2022-08-24 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of substrate-free DNClpX.ClpP
Nat Commun, 2023
|
|
8E91
 
 | |
8E8Q
 
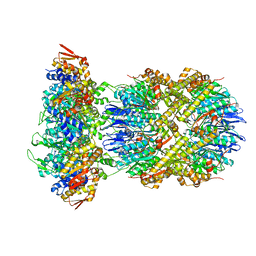 | | Cryo-EM structure of substrate-free DNClpX.ClpP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | Authors: | Ghanbarpour, A, Cohen, S, Davis, J.H, Sauer, R.T. | | Deposit date: | 2022-08-25 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structure of substrate-free DNClpX.ClpP
Nat Commun, 2023
|
|
3LGU
 
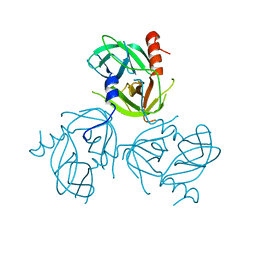 | | Y162A mutant of the DegS-deltaPDZ protease | | Descriptor: | Protease degS | | Authors: | Sohn, J, Grant, R.A, Sauer, R.T. | | Deposit date: | 2010-01-21 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Allostery is an intrinsic property of the protease domain of DegS: implications for enzyme function and evolution.
J.Biol.Chem., 285, 2010
|
|
2QAS
 
 | |
4WNQ
 
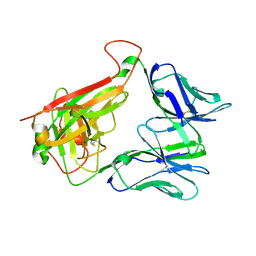 | | THE MOLECULAR BASES OF DELTA/ALPHA-BETA T-CELL MEDIATED ANTIGEN RECOGNITION | | Descriptor: | TCR Variable Beta 2 (TRBV20) chain and TCR constant Beta chain, TCR Variable Delta 1 chain and TCR constant Alpha chain | | Authors: | Pellicci, D.G, Uldrich, A.P, Le Nours, J, Ross, F, Chabrol, E, Eckle, S.B.G, de Boer, R, Lim, R.T, McPherson, K, Besra, G, Howell, A.R, Moretta, L, McCluskey, J, Heemskerk, M.H.M, Gras, S, Rossjohn, J, Godfrey, D.I. | | Deposit date: | 2014-10-14 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular bases of delta / alpha beta T cell-mediated antigen recognition.
J.Exp.Med., 211, 2014
|
|
4WO4
 
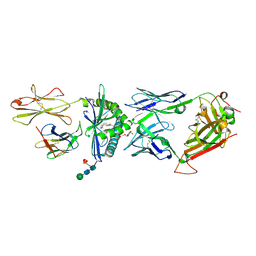 | | The molecular bases of Delta/Alpha beta T cell-mediated antigen recognition. | | Descriptor: | (15Z)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]tetracos-15-enamide, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pellicci, D.G, Uldrich, A.P, Le Nours, J, Ross, F, Chabrol, E, Eckle, S.B.G, de Boer, R, Lim, R.T, McPherson, K, Besra, G, Howell, A.R, Moretta, L, McCluskey, J, Heemskerk, M.H.M, Gras, S, Rossjohn, J, Godfrey, D.I. | | Deposit date: | 2014-10-15 | | Release date: | 2014-11-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The molecular bases of delta / alpha beta T cell-mediated antigen recognition.
J.Exp.Med., 211, 2014
|
|
2QF3
 
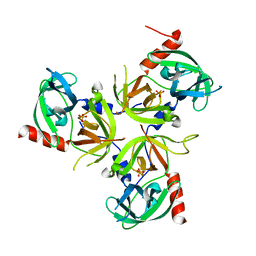 | | Structure of the delta PDZ truncation of the DegS protease | | Descriptor: | PHOSPHATE ION, Protease degS | | Authors: | Sohn, J, Grant, R.A, Sauer, R.T. | | Deposit date: | 2007-06-26 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Allosteric activation of DegS, a stress sensor PDZ protease.
Cell(Cambridge,Mass.), 131, 2007
|
|
3LGY
 
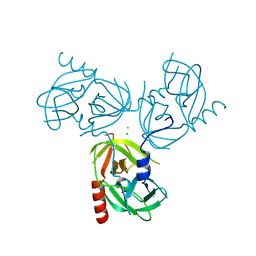 | | R178A mutant of the DegS-deltaPDZ protease | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Protease degS | | Authors: | Sohn, J, Grant, R.A, Sauer, R.T. | | Deposit date: | 2010-01-21 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allostery is an intrinsic property of the protease domain of DegS: implications for enzyme function and evolution.
J.Biol.Chem., 285, 2010
|
|
3LQX
 
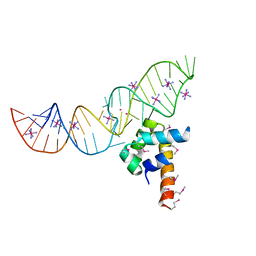 | | SRP ribonucleoprotein core complexed with cobalt hexammine | | Descriptor: | CHLORIDE ION, COBALT HEXAMMINE(III), POTASSIUM ION, ... | | Authors: | Batey, R.T. | | Deposit date: | 2010-02-10 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and Energetic Analysis of Metal Ions Essential to SRP Signal Recognition Domain Assembly
Biochemistry, 41, 2002
|
|
8ET3
 
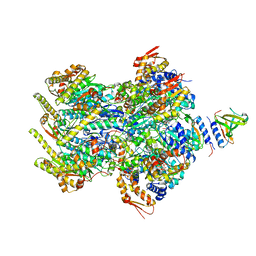 | | Cryo-EM structure of a delivery complex containing the SspB adaptor, an ssrA-tagged substrate, and the AAA+ ClpXP protease | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Ghanbarpour, A, Fei, X, Davis, J.H, Sauer, R.T. | | Deposit date: | 2022-10-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The SspB adaptor drives structural changes in the AAA+ ClpXP protease during ssrA-tagged substrate delivery.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ESX
 
 | | HIV protease in complex with benzoxaborolone analog of darunavir | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Windsor, I.W, Graham, B.J, Raines, R.T. | | Deposit date: | 2022-10-15 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Inhibition of HIV-1 Protease by a Boronic Acid with High Oxidative Stability.
Acs Med.Chem.Lett., 14, 2023
|
|
