4WTB
 
 | | BthTX-I, a svPLA2s-like toxin, complexed with zinc ions | | Descriptor: | Basic phospholipase A2 homolog bothropstoxin-1, CHLORIDE ION, SULFATE ION, ... | | Authors: | Borges, R.J, Fontes, M.R.M. | | Deposit date: | 2014-10-29 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Functional and structural studies of a Phospholipase A2-like protein complexed to zinc ions: Insights on its myotoxicity and inhibition mechanism.
Biochim. Biophys. Acta, 1861, 2017
|
|
4MPE
 
 | | Crystal structure of pyruvate dehydrogenase kinase isoform 2 in complex with inhibitor PS8 | | Descriptor: | 4-[(5-hydroxy-1,3-dihydro-2H-isoindol-2-yl)sulfonyl]benzene-1,3-diol, L(+)-TARTARIC ACID, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 2, ... | | Authors: | Gui, W.J, Tso, S.C, Chuang, J.L, Wu, C.Y, Qi, X, Tambar, U.K, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2013-09-12 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Structure-guided Development of Specific Pyruvate Dehydrogenase Kinase Inhibitors Targeting the ATP-binding Pocket.
J.Biol.Chem., 289, 2014
|
|
6NCG
 
 | | Crystal Structure of Human Vaccinia-related kinase 2 (VRK-2) bound to pyridin-benzenesulfonamide inhibitor | | Descriptor: | 4-[6-amino-5-(3,5-difluoro-4-hydroxyphenyl)pyridin-3-yl]benzene-1-sulfonamide, SULFATE ION, Serine/threonine-protein kinase VRK2 | | Authors: | dos Reis, C.V, Chiodi, C.G, de Souza, G.P, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Counago, R.M, Massirer, K.B, Elkins, J.M, Arruda, P, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-11 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Human Vaccinia-related kinase 2 (VRK-2) bound to pyridin-benzenesulfonamide inhibitor
To Be Published
|
|
6NNR
 
 | | high-resolution structure of wild-type E. coli thymidylate synthase | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-deoxy-5'-uridylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Stroud, R.M, Finer-Moore, J. | | Deposit date: | 2019-01-15 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Mg2+ binds to the surface of thymidylate synthase and affects hydride transfer at the interior active site.
J. Am. Chem. Soc., 135, 2013
|
|
4NJK
 
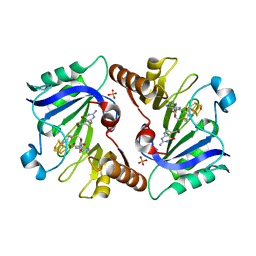 | | Crystal Structure of QueE from Burkholderia multivorans in complex with AdoMet, 7-carboxy-7-deazaguanine, and Mg2+ | | Descriptor: | 2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carboxylic acid, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Bruender, N.A, Young, A.P, McCarty, R.M, Bandarian, V, Drennan, C.L. | | Deposit date: | 2013-11-10 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.911 Å) | | Cite: | Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism.
Nat.Chem.Biol., 10, 2014
|
|
4X9E
 
 | | DEOXYGUANOSINETRIPHOSPHATE TRIPHOSPHOHYDROLASE from Escherichia coli with two DNA effector molecules | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, MAGNESIUM ION, RNA (5'-R(P*CP*CP*C)-3') | | Authors: | Singh, D, Gawel, D, Itsko, M, Krahn, J.M, London, R.E, Schaaper, R.M. | | Deposit date: | 2014-12-11 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Escherichia coli dGTP Triphosphohydrolase: A HEXAMERIC ENZYME WITH DNA EFFECTOR MOLECULES.
J.Biol.Chem., 290, 2015
|
|
4WTX
 
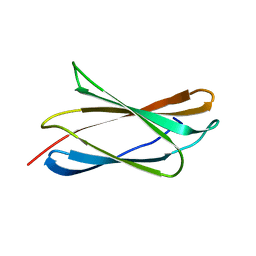 | | Crystal structure of the fourth FnIII domain of integrin beta4 | | Descriptor: | Integrin beta-4 | | Authors: | Alonso-Garcia, N, Urien, H, Buey, R.M, de Pereda, J.M. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Combination of X-ray crystallography, SAXS and DEER to obtain the structure of the FnIII-3,4 domains of integrin alpha6beta4
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7XAY
 
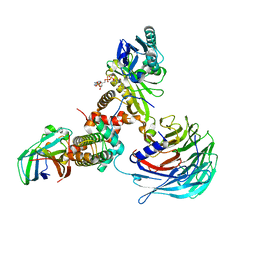 | | Crystal structure of Hat1-Hat2-Asf1-H3-H4 | | Descriptor: | COENZYME A, Histone H3, Histone H4, ... | | Authors: | Yue, Y, Yang, W.S, Xu, R.M. | | Deposit date: | 2022-03-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Topography of histone H3-H4 interaction with the Hat1-Hat2 acetyltransferase complex.
Genes Dev., 36, 2022
|
|
4OO9
 
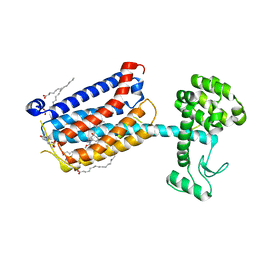 | | Structure of the human class C GPCR metabotropic glutamate receptor 5 transmembrane domain in complex with the negative allosteric modulator mavoglurant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mavoglurant, Metabotropic glutamate receptor 5, ... | | Authors: | Dore, A.S, Okrasa, K, Patel, J.C, Serrano-Vega, M, Bennett, K, Cooke, R.M, Errey, J.C, Jazayeri, A, Khan, S, Tehan, B, Weir, M, Wiggin, G.R, Marshall, F.H. | | Deposit date: | 2014-01-31 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain.
Nature, 511, 2014
|
|
1BFR
 
 | | IRON STORAGE AND ELECTRON TRANSPORT | | Descriptor: | BACTERIOFERRITIN, MANGANESE (II) ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dautant, A, Yariv, J, Meyer, J.B, Precigoux, G, Sweet, R.M, Frolow, F, Kalb(Gilboa), A.J. | | Deposit date: | 1994-12-16 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structure of a monoclinic crystal from of cyctochrome b1 (Bacterioferritin) from E. coli.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
4OX5
 
 | | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Hoyland, C.N, Aldridge, C, Cleverley, R.M, Sidiq, K, Duchene, M.C, Daniel, R.A, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-02-04 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition.
Structure, 22, 2014
|
|
4OXD
 
 | | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition | | Descriptor: | CHLORIDE ION, LYSINE, LdcB LD-carboxypeptidase, ... | | Authors: | Hoyland, C.N, Aldridge, C, Cleverley, R.M, Sidiq, K, Duchene, M.C, Daniel, R.A, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-02-05 | | Release date: | 2014-05-21 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition.
Structure, 22, 2014
|
|
2I5J
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with DHBNH, an RNASE H inhibitor | | Descriptor: | (E)-3,4-DIHYDROXY-N'-[(2-METHOXYNAPHTHALEN-1-YL)METHYLENE]BENZOHYDRAZIDE, MAGNESIUM ION, Reverse transcriptase/ribonuclease H P51 subunit, ... | | Authors: | Himmel, D.M, Sarafianos, S.G, Knight, J.L, Levy, R.M, Arnold, E. | | Deposit date: | 2006-08-24 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | HIV-1 reverse transcriptase structure with RNase H inhibitor dihydroxy benzoyl naphthyl hydrazone bound at a novel site.
Acs Chem.Biol., 1, 2006
|
|
4OAA
 
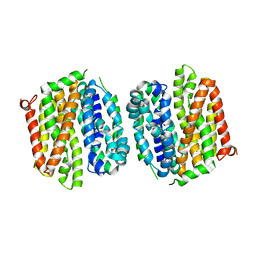 | | Crystal structure of E. coli lactose permease G46W,G262W bound to sugar | | Descriptor: | Lactose/galactose transporter, beta-D-galactopyranose-(1-1)-1-thio-beta-D-galactopyranose | | Authors: | Kumar, H, Kasho, V, Smirnova, I, Finer-Moore, J, Kaback, H.R, Stroud, R.M. | | Deposit date: | 2014-01-03 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of sugar-bound LacY.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1DTW
 
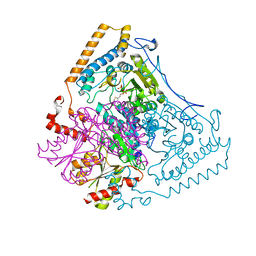 | | HUMAN BRANCHED-CHAIN ALPHA-KETO ACID DEHYDROGENASE | | Descriptor: | BRANCHED-CHAIN ALPHA-KETO ACID DEHYDROGENASE ALPHA SUBUNIT, BRANCHED-CHAIN ALPHA-KETO ACID DEHYDROGENASE BETA SUBUNIT, MAGNESIUM ION, ... | | Authors: | AEvarsson, A, Chuang, J.L, Wynn, R.M, Turley, S, Chuang, D.T, Hol, W.G.J. | | Deposit date: | 2000-01-13 | | Release date: | 2000-03-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human branched-chain alpha-ketoacid dehydrogenase and the molecular basis of multienzyme complex deficiency in maple syrup urine disease.
Structure Fold.Des., 8, 2000
|
|
6NBS
 
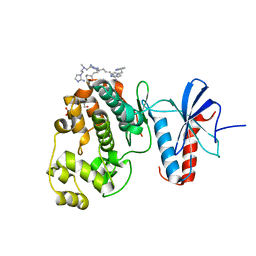 | | WT ERK2 with compound 2507-8 | | Descriptor: | (5S)-5-benzyl-4,5-dihydro-1H-imidazol-2-amine, GLYCEROL, Mitogen-activated protein kinase 1, ... | | Authors: | Sammons, R.M, Perry, N.A, Cho, E.J, Kaoud, T.S, Zamora-Olivares, D.P, Piserchio, A, Houghten, R.A, Giulianotti, M, Li, Y, Debevec, G, Gurevich, V.V, Ghose, R, Iverson, T.M, Dalby, K.N. | | Deposit date: | 2018-12-10 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Novel Class of Common Docking Domain Inhibitors That Prevent ERK2 Activation and Substrate Phosphorylation.
Acs Chem.Biol., 14, 2019
|
|
1DZ4
 
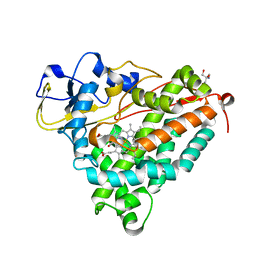 | | ferric p450cam from pseudomonas putida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYTOCHROME P450-CAM, ... | | Authors: | Schlichting, I, Berendzen, J, Chu, K, Stock, A.M, Maves, S.A, Benson, D.E, Sweet, R.M, Ringe, D, Petsko, G.A, Sligar, S.G. | | Deposit date: | 2000-02-16 | | Release date: | 2000-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Cytochrome P450Cam at Atomic Resolution
Science, 287, 2000
|
|
2II4
 
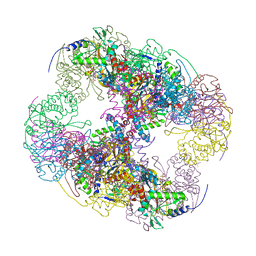 | | Crystal structure of a cubic core of the dihydrolipoamide acyltransferase (E2b) component in the branched-chain alpha-ketoacid dehydrogenase complex (BCKDC), Coenzyme A-bound form | | Descriptor: | CHLORIDE ION, COENZYME A, Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Brautigam, C.A, Custorio, M, Chuang, D.T. | | Deposit date: | 2006-09-27 | | Release date: | 2006-12-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | A synchronized substrate-gating mechanism revealed by cubic-core structure of the bovine branched-chain alpha-ketoacid dehydrogenase complex.
Embo J., 25, 2006
|
|
2IDO
 
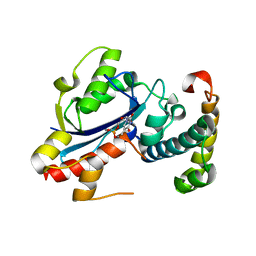 | | Structure of the E. coli Pol III epsilon-Hot proofreading complex | | Descriptor: | 1,2-ETHANEDIOL, DNA polymerase III epsilon subunit, Hot protein, ... | | Authors: | Kirby, T.W, Harvey, S, DeRose, E.F, Chalov, S, Chikova, A.K, Perrino, F.W, Schaaper, R.M, London, R.E, Pedersen, L.C. | | Deposit date: | 2006-09-15 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Escherichia coli DNA polymerase III epsilon-HOT proofreading complex.
J.Biol.Chem., 281, 2006
|
|
1XAE
 
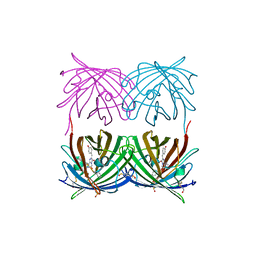 | | Crystal structure of wild type yellow fluorescent protein zFP538 from Zoanthus | | Descriptor: | BETA-MERCAPTOETHANOL, fluorescent protein FP538 | | Authors: | Remington, S.J, Wachter, R.M, Yarbrough, D.K, Branchaud, B, Anderson, D.C, Kallio, K, Lukyanov, K.A. | | Deposit date: | 2004-08-25 | | Release date: | 2005-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | zFP538, a yellow-fluorescent protein from Zoanthus, contains a novel three-ring chromophore.
Biochemistry, 44, 2005
|
|
2II3
 
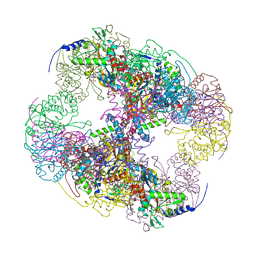 | | Crystal structure of a cubic core of the dihydrolipoamide acyltransferase (E2b) component in the branched-chain alpha-ketoacid dehydrogenase complex (BCKDC), Oxidized Coenzyme A-bound form | | Descriptor: | ACETATE ION, CHLORIDE ION, Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Brautigam, C.A, Custorio, M, Chuang, D.T. | | Deposit date: | 2006-09-27 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A synchronized substrate-gating mechanism revealed by cubic-core structure of the bovine branched-chain alpha-ketoacid dehydrogenase complex.
Embo J., 25, 2006
|
|
1DXC
 
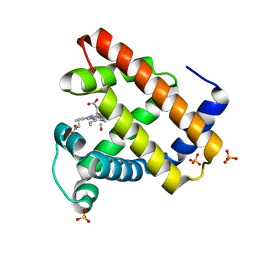 | | CO complex of Myoglobin Mb-YQR at 100K | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Brunori, M, Vallone, B, Cutruzzola, F, Travaglini-Allocatelli, C, Berendzen, J, Chu, K, Sweet, R.M, Schlichting, I. | | Deposit date: | 2000-01-03 | | Release date: | 2000-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Role of Cavities in Protein Dynamics: Crystal Structure of a Novel Photolytic Intermediate of Myoglobin
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
6O1G
 
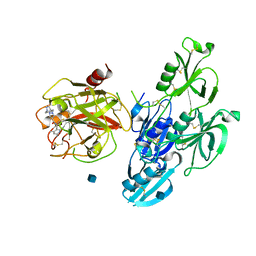 | | Full length human plasma kallikrein with inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-[(6-amino-2,4-dimethylpyridin-3-yl)methyl]-1-({4-[(1H-pyrazol-1-yl)methyl]phenyl}methyl)-1H-pyrazole-4-carboxamide, Plasma kallikrein | | Authors: | Partridge, J.R, Choy, R.M. | | Deposit date: | 2019-02-19 | | Release date: | 2019-03-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of full-length plasma kallikrein bound to highly specific inhibitors describe a new mode of targeted inhibition.
J.Struct.Biol., 206, 2019
|
|
1DZC
 
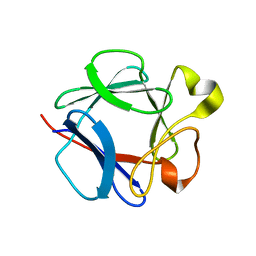 | | High resolution structure of acidic fibroblast growth factor. Mutant FGF-4-ALA-(24-154), 24 NMR structures | | Descriptor: | FIBROBLAST GROWTH FACTOR 1 | | Authors: | Lozano, R.M, Pineda-Lucena, A, Gonzalez, C, Jimenez, M.A, Cuevas, P, Redondo-Horcajo, M, Sanz, J.M, Rico, M, Gimenez-Gallego, G. | | Deposit date: | 2000-02-24 | | Release date: | 2000-03-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H-NMR Structural Characterization of a Non Mitogenic, Vasodilatory, Ischemia-Protector and Neuromodulatory Acidic Fibroblast Growth Factor
Biochemistry, 39, 2000
|
|
1XEQ
 
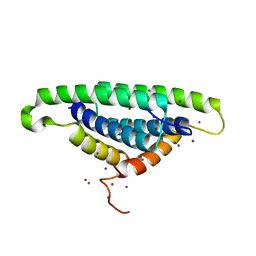 | | Crystal tructure of RNA binding domain of influenza B virus non-structural protein | | Descriptor: | BROMIDE ION, Nonstructural protein NS1 | | Authors: | Khan, J.A, Yin, C, Krug, R.M, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-09-11 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conserved surface features form the double-stranded RNA binding site of non-structural protein 1 (NS1) from influenza A and B viruses.
J.Biol.Chem., 282, 2007
|
|
