1QIQ
 
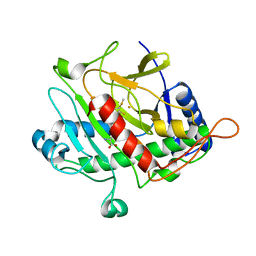 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (ACmC Fe COMPLEX) | | Descriptor: | FE (III) ION, ISOPENICILLIN N SYNTHASE, N-[N-[2-AMINO-6-OXO-HEXANOIC ACID-6-YL]CYSTEINYL]-S-METHYLCYSTEINE, ... | | Authors: | Rutledge, P.J, Clifton, I.J, Burzlaff, N.I, Roach, P.L, Adlington, R.M, Baldwin, J.E. | | Deposit date: | 1999-06-15 | | Release date: | 2000-06-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Reaction Cycle of Isopenicillin N Synthase Observed by X-Ray Diffraction.
Nature, 401, 1999
|
|
1QJF
 
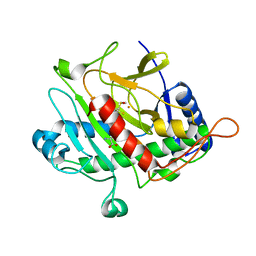 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (Monocyclic Sulfoxide - Fe COMPLEX) | | Descriptor: | 1-[(1S)-CARBOXY-2-(METHYLSULFINYL)ETHYL]-(3R)-[(5S)-5-AMINO-5-CARBOXYPENTANAMIDO]-(4R)-SULFANYLAZETIDIN-2-ONE, FE (II) ION, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Rutledge, P.J, Clifton, I.J, Burzlaff, N.I, Roach, P.L, Adlington, R.M, Baldwin, J.E. | | Deposit date: | 1999-06-23 | | Release date: | 2000-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Reaction Cycle of Isopenicillin N Synthase Observed by X-Ray Diffraction
Nature, 401, 1999
|
|
1QJE
 
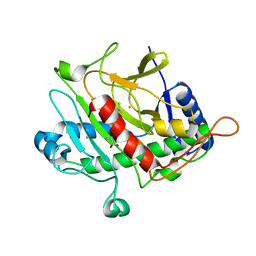 | | Isopenicillin N synthase from Aspergillus nidulans (IP1 - Fe complex) | | Descriptor: | FE (II) ION, ISOPENICILLIN N, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Burzlaff, N.I, Clifton, I.J, Rutledge, P.J, Roach, P.L, Adlington, R.M, Baldwin, J.E. | | Deposit date: | 1999-06-23 | | Release date: | 2000-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Reaction Cycle of Isopenicillin N Synthase Observed by X-Ray Diffraction
Nature, 401, 1999
|
|
1OSF
 
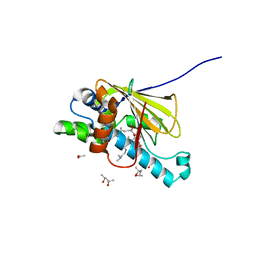 | | Human Hsp90 in complex with 17-desmethoxy-17-N,N-Dimethylaminoethylamino-Geldanamycin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 17-DESMETHOXY-17-N,N-DIMETHYLAMINOETHYLAMINO-GELDANAMYCIN, ACETIC ACID, ... | | Authors: | Jez, J.M, Chen, J.C.-H, Rastelli, G, Stroud, R.M, Santi, D.V. | | Deposit date: | 2003-03-19 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Molecular Modeling of 17-DMAG in Complex with Human Hsp90
Chem.Biol., 10, 2003
|
|
1I2B
 
 | | CRYSTAL STRUCTURE OF MUTANT T145A SQD1 PROTEIN COMPLEX WITH NAD AND UDP-SULFOQUINOVOSE/UDP-GLUCOSE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, SULFOLIPID BIOSYNTHESIS PROTEIN SQD1, ... | | Authors: | Theisen, M.J, Sanda, S.L, Ginell, S.L, Benning, C, Garavito, R.M. | | Deposit date: | 2001-02-07 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization of the Active Site of UDP-sulfoquinovose Synthase: Formation of the Sulfonic Acid Product in the Crystalline State
To be Published
|
|
2RKA
 
 | | The Structure of rat cytosolic PEPCK in complex with phosphoglycolate | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, MANGANESE (II) ION, Phosphoenolpyruvate carboxykinase, ... | | Authors: | Sullivan, S.M, Stiffin, R.M, Carlson, G.M, Holyoak, T. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Differential Inhibition of Cytosolic PEPCK by Substrate Analogues. Kinetic and Structural Characterization of Inhibitor Recognition.
Biochemistry, 47, 2008
|
|
2Q9Z
 
 | | Trichodiene synthase: Complex with inorganic pyrophosphate resulting from the reaction with 2-fluorofarnesyl diphosphate | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Vedula, L.S, Zhao, Y, Coates, R.M, Koyama, T, Cane, D.E, Christianson, D.W. | | Deposit date: | 2007-06-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Exploring biosynthetic diversity with trichodiene synthase.
Arch.Biochem.Biophys., 466, 2007
|
|
1PWX
 
 | | Crystal structure of the haloalcohol dehalogenase HheC complexed with bromide | | Descriptor: | BROMIDE ION, halohydrin dehalogenase | | Authors: | de Jong, R.M, Tiesinga, J.J.W, Rozeboom, H.J, Kalk, K.H, Tang, L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2003-07-02 | | Release date: | 2003-10-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Mechanism of a Bacterial Haloalcohol Dehalogenase: a new variation of the short-chain dehydrogenase/reductase fold without an NAD(P)H binding site
EMBO J., 22, 2003
|
|
1HD9
 
 | | The Bowman-Birk Inhibitor Reactive Site Loop Sequence Represents an Independent Structural Beta-Hairpin Motif | | Descriptor: | BOWMAN-BIRK INHIBITOR DERIVED PEPTIDE | | Authors: | Brauer, A.B.E, Kelly, G, McBride, J.D, Cooke, R.M, Matthews, S.J, Leatherbarrow, R.J. | | Deposit date: | 2000-11-13 | | Release date: | 2001-03-29 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The Bowman-Birk Inhibitor Reactive Site Loop Sequence Represents an Independent Structural Beta-Hairpin Motif
J.Mol.Biol., 306, 2001
|
|
1PRY
 
 | | Structure Determination of Fibrillarin Homologue From Hyperthermophilic Archaeon Pyrococcus furiosus (Pfu-65527) | | Descriptor: | Fibrillarin-like pre-rRNA processing protein | | Authors: | Deng, L, Starostina, N.G, Liu, Z.-J, Rose, J.P, Terns, R.M, Terns, M.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-06-20 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure determination of fibrillarin from the hyperthermophilic archaeon Pyrococcus furiosus
Biochem.Biophys.Res.Commun., 315, 2004
|
|
1I24
 
 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE WILD-TYPE PROTEIN SQD1, WITH NAD AND UDP-GLUCOSE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, SULFOLIPID BIOSYNTHESIS PROTEIN SQD1, ... | | Authors: | Theisen, M.J, Sanda, S.L, Ginell, S.L, Benning, C, Garavito, R.M. | | Deposit date: | 2001-02-05 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Characterization of the Active Site of Udp-Sulfoquinovose Synthase: Formation of the Sulfonic Acid Product in the Crystalline State
To be Published
|
|
1PN3
 
 | | Crystal Structure of TDP-epi-Vancosaminyltransferase GtfA in complexes with TDP and the acceptor substrate DVV. | | Descriptor: | DESVANCOSAMINYL VANCOMYCIN, GLYCOSYLTRANSFERASE GTFA, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Mulichak, A.M, Losey, H.C, Lu, W, Wawrzak, Z, Walsh, C.T, Garavito, R.M. | | Deposit date: | 2003-06-12 | | Release date: | 2003-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Tdp-Epi-Vancosaminyltransferase Gtfa from the Chloroeremomycin Biosynthetic Pathway.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1HRJ
 
 | | HUMAN RANTES, NMR, 13 STRUCTURES | | Descriptor: | HUMAN REGULATED UPON ACTIVATION NORMAL T-CELL EXPRESSED AND SECRETED | | Authors: | Chung, C, Cooke, R.M, Proudfoot, A.E.I, Wells, T.N.C. | | Deposit date: | 1995-08-18 | | Release date: | 1996-10-14 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of RANTES.
Biochemistry, 34, 1995
|
|
2Q9Y
 
 | | Trichodiene synthase: Complex with Mg, inorganic pyrophosphate, and benzyl triethyl ammonium cation | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium, ... | | Authors: | Vedula, L.S, Zhao, Y, Coates, R.M, Koyama, T, Cane, D.E, Christianson, D.W. | | Deposit date: | 2007-06-14 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Exploring biosynthetic diversity with trichodiene synthase.
Arch.Biochem.Biophys., 466, 2007
|
|
1FFH
 
 | | N AND GTPASE DOMAINS OF THE SIGNAL SEQUENCE RECOGNITION PROTEIN FFH FROM THERMUS AQUATICUS | | Descriptor: | FFH, MAGNESIUM ION | | Authors: | Freymann, D.M, Keenan, R.J, Stroud, R.M, Walter, P. | | Deposit date: | 1996-12-30 | | Release date: | 1997-12-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the conserved GTPase domain of the signal recognition particle.
Nature, 385, 1997
|
|
1HP9
 
 | | kappa-Hefutoxins: a novel Class of Potassium Channel Toxins from Scorpion venom | | Descriptor: | kappa-hefutoxin 1 | | Authors: | Srinivasan, K.N, Sivaraja, V, Huys, I, Sasaki, T, Cheng, B, Kumar, T.K.S, Sato, K, Tytgat, J, Yu, C, Brian Chia, C.S, Ranganathan, S, Bowie, J.H, Kini, R.M, Gopalakrishnakone, P. | | Deposit date: | 2000-12-12 | | Release date: | 2002-08-28 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | kappa-Hefutoxin1, a novel toxin from the scorpion Heterometrus fulvipes with unique structure and function. Importance of the functional diad in potassium channel selectivity.
J.Biol.Chem., 277, 2002
|
|
1HV8
 
 | |
7LUH
 
 | |
7LUJ
 
 | |
2RKE
 
 | | The Structure of rat cytosolic PEPCK in complex with sulfoacetate. | | Descriptor: | MANGANESE (II) ION, Phosphoenolpyruvate carboxykinase, cytosolic [GTP], ... | | Authors: | Sullivan, S.M, Stiffin, R.M, Carlson, G.M, Holyoak, T. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Differential Inhibition of Cytosolic PEPCK by Substrate Analogues. Kinetic and Structural Characterization of Inhibitor Recognition.
Biochemistry, 47, 2008
|
|
2SAM
 
 | | STRUCTURE OF THE PROTEASE FROM SIMIAN IMMUNODEFICIENCY VIRUS: COMPLEX WITH AN IRREVERSIBLE NON-PEPTIDE INHIBITOR | | Descriptor: | 3-(4-NITRO-PHENOXY)-PROPAN-1-OL, SIV PROTEASE | | Authors: | Rose, R.B, Rose, J.R, Salto, R, Craik, C.S, Stroud, R.M. | | Deposit date: | 1994-07-08 | | Release date: | 1994-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the protease from simian immunodeficiency virus: complex with an irreversible nonpeptide inhibitor.
Biochemistry, 32, 1993
|
|
1S8J
 
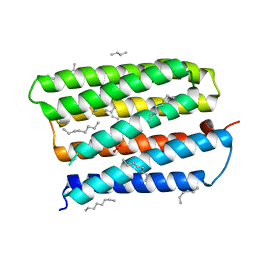 | | Nitrate-bound D85S mutant of bacteriorhodopsin | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin precursor, NITRATE ION, ... | | Authors: | Facciotti, M.T, Cheung, V.S, Lunde, C.S, Rouhani, S, Baliga, N.S, Glaeser, R.M. | | Deposit date: | 2004-02-02 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Specificity of anion binding in the substrate pocket of bacteriorhodopsin.
Biochemistry, 43, 2004
|
|
2RK7
 
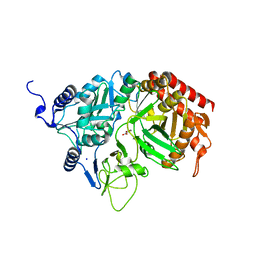 | | The Structure of rat cytosolic PEPCK in complex with oxalate | | Descriptor: | MANGANESE (II) ION, OXALATE ION, Phosphoenolpyruvate carboxykinase, ... | | Authors: | Sullivan, S.M, Stiffin, R.M, Carlson, G.M, Holyoak, T. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential Inhibition of Cytosolic PEPCK by Substrate Analogues. Kinetic and Structural Characterization of Inhibitor Recognition.
Biochemistry, 47, 2008
|
|
1P0R
 
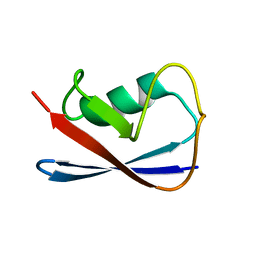 | | Solution Structure of UBL5 a human Ubiquitin-Like Protein | | Descriptor: | ubiquitin-like 5 | | Authors: | McNally, T, Huang, Q, Janis, R.S, Liu, Z, Olejniczak, E.T, Reilly, R.M. | | Deposit date: | 2003-04-10 | | Release date: | 2003-10-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of UBL5, a novel ubiquitin-like modifier
Protein Sci., 12, 2003
|
|
1OSH
 
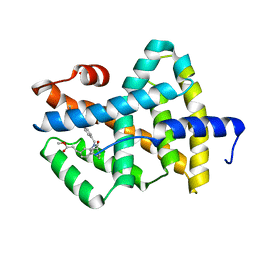 | | A Chemical, Genetic, and Structural Analysis of the nuclear bile acid receptor FXR | | Descriptor: | Bile acid receptor, METHYL 3-{3-[(CYCLOHEXYLCARBONYL){[4'-(DIMETHYLAMINO)BIPHENYL-4-YL]METHYL}AMINO]PHENYL}ACRYLATE | | Authors: | Downes, M, Verdecia, M.A, Roecker, A.J, Hughes, R, Hogenesch, J.B, Kast-Woelbern, H.R, Bowman, M.E, Ferrer, J.-L, Anisfeld, A.M, Edwards, P.A, Rosenfeld, J.M, Alvarez, J.G.A, Noel, J.P, Nicolaou, K.C, Evans, R.M. | | Deposit date: | 2003-03-19 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR
Mol.Cell, 11, 2003
|
|
