7GIS
 
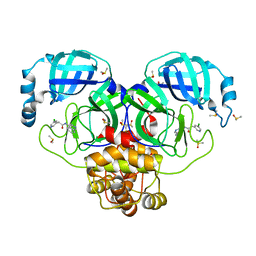 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with RAL-THA-2d450e86-30 (Mpro-P0126) | | Descriptor: | 2-(5-chloropyridin-3-yl)-N-(isoquinolin-4-yl)acetamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.838 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GJL
 
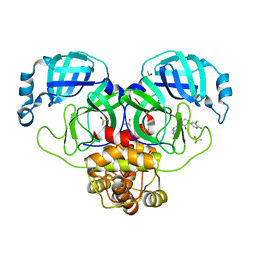 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-UNI-3735e77e-1 (Mpro-P0600) | | Descriptor: | (3R)-3-(3,4-dichlorophenyl)-1-(isoquinolin-4-yl)piperidin-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
1Q1T
 
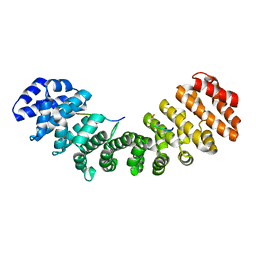 | | Mouse Importin alpha: non-phosphorylated SV40 CN peptide complex | | Descriptor: | Importin alpha-2 subunit, Large T antigen | | Authors: | Fontes, M.R.M, Teh, T, Toth, G, John, A, Pavo, I, Jans, D.A, Kobe, B. | | Deposit date: | 2003-07-22 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Role of flanking sequences and phosphorylation in the recognition of the simian-virus-40 large T-antigen nuclear localization sequences by importin-alpha
Biochem.J., 375, 2003
|
|
7LSO
 
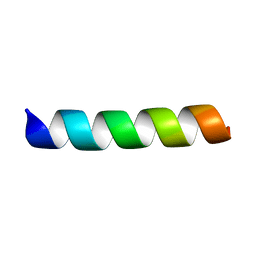 | | L-Phenylseptin | | Descriptor: | L-Phenylseptin peptide | | Authors: | Munhoz, V.H, Verly, R.M. | | Deposit date: | 2021-02-18 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of L-Phenylseptin in DPC-d38.
To Be Published
|
|
7LSP
 
 | |
1Q1S
 
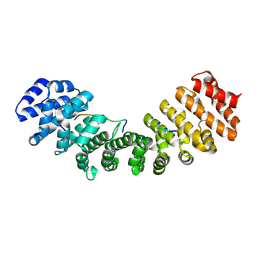 | | Mouse Importin alpha- phosphorylated SV40 CN peptide complex | | Descriptor: | Importin alpha-2 subunit, Large T antigen | | Authors: | Fontes, M.R.M, Teh, T, Toth, G, John, A, Pavo, I, Jans, D.A, Kobe, B. | | Deposit date: | 2003-07-22 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of flanking sequences and phosphorylation in the recognition of the simian-virus-40 large T-antigen nuclear localization sequences by importin-alpha
Biochem.J., 375, 2003
|
|
7KRA
 
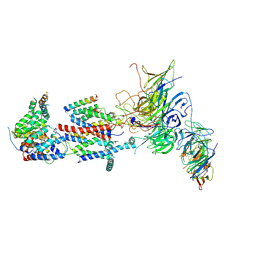 | | Cryo-EM structure of Saccharomyces cerevisiae ER membrane protein complex bound to Fab-DH4 in lipid nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ER membrane protein complex subunit 2, ... | | Authors: | Miller-Vedam, L.E, Schirle Oakdale, N.S, Braeuning, B, Boydston, E.A, Sevillano, N, Popova, K.D, Bonnar, J.L, Shurtleff, M.J, Prabu, J.R, Stroud, R.M, Craik, C.S, Schulman, B.A, Weissman, J.S, Frost, A. | | Deposit date: | 2020-11-19 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
7KUU
 
 | |
7MTO
 
 | |
7MN3
 
 | | Homotarsinin monomer - Htr-M | | Descriptor: | Homotarsinin | | Authors: | Verly, R.M. | | Deposit date: | 2021-04-30 | | Release date: | 2021-07-14 | | Method: | SOLUTION NMR | | Cite: | Structure and membrane interactions of the homodimeric antibiotic peptide homotarsinin.
Sci Rep, 7, 2017
|
|
1PU0
 
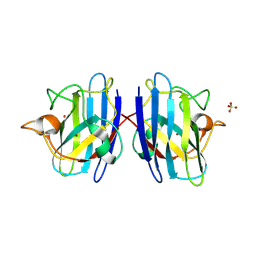 | | Structure of Human Cu,Zn Superoxide Dismutase | | Descriptor: | COPPER (I) ION, SULFATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | DiDonato, M, Craig, L, Huff, M.E, Thayer, M.M, Cardoso, R.M.F, Kassmann, C.J, Lo, T.P, Bruns, C.K, Powers, E.T, Kelly, J.W, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | ALS Mutants of Human Superoxide Dismutase Form Fibrous Aggregates Via Framework Destabilization
J.Mol.Biol., 332, 2003
|
|
4HU4
 
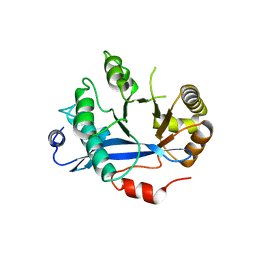 | | Crystal structure of EAL domain of the E. coli DosP - dimeric form | | Descriptor: | Oxygen sensor protein DosP | | Authors: | Tarnawski, M, Barends, T.R.M, Hartmann, E, Schlichting, I. | | Deposit date: | 2012-11-02 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the catalytic EAL domain of the Escherichia coli direct oxygen sensor.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HWY
 
 | | Trypanosoma brucei procathepsin B solved from 40 fs free-electron laser pulse data by serial femtosecond X-ray crystallography | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cysteine peptidase C (CPC), beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Redecke, L, Nass, K, DePonte, D.P, White, T.A, Rehders, D, Barty, A, Stellato, F, Liang, M, Barends, T.R.M, Boutet, S, Williams, G.W, Messerschmidt, M, Seibert, M.M, Aquila, A, Arnlund, D, Bajt, S, Barth, T, Bogan, M.J, Caleman, C, Chao, T.-C, Doak, R.B, Fleckenstein, H, Frank, M, Fromme, R, Galli, L, Grotjohann, I, Hunter, M.S, Johansson, L.C, Kassemeyer, S, Katona, G, Kirian, R.A, Koopmann, R, Kupitz, C, Lomb, L, Martin, A.V, Mogk, S, Neutze, R, Shoemann, R.L, Steinbrener, J, Timneanu, N, Wang, D, Weierstall, U, Zatsepin, N.A, Spence, J.C.H, Fromme, P, Schlichting, I, Duszenko, M, Betzel, C, Chapman, H. | | Deposit date: | 2012-11-09 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser.
Science, 339, 2013
|
|
4HU3
 
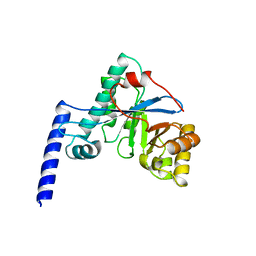 | | Crystal structure of EAL domain of the E. coli DosP - monomeric form | | Descriptor: | Oxygen sensor protein DosP | | Authors: | Tarnawski, M, Barends, T.R.M, Hartmann, E, Schlichting, I. | | Deposit date: | 2012-11-02 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | Structures of the catalytic EAL domain of the Escherichia coli direct oxygen sensor.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4FHE
 
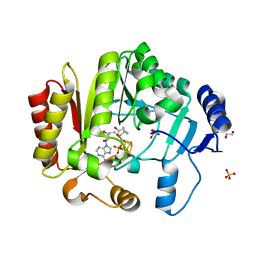 | | Spore photoproduct lyase C140A mutant | | Descriptor: | 1,2-ETHANEDIOL, IRON/SULFUR CLUSTER, SULFATE ION, ... | | Authors: | Benjdia, A, Heil, K, Barends, T.R.M, Carell, T, Schlichting, I. | | Deposit date: | 2012-06-06 | | Release date: | 2012-07-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into recognition and repair of UV-DNA damage by Spore Photoproduct Lyase, a radical SAM enzyme.
Nucleic Acids Res., 40, 2012
|
|
4FGE
 
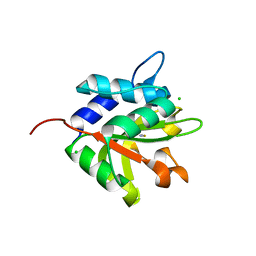 | | Structure of the effector protein Tse1 from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, GLYCEROL, THIOCYANATE ION, ... | | Authors: | Benz, J, Sendlmeier, C, Barends, T.R.M, Meinhart, A. | | Deposit date: | 2012-06-04 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights into the Effector - Immunity System Tse1/Tsi1 from Pseudomonas aeruginosa.
Plos One, 7, 2012
|
|
4FHD
 
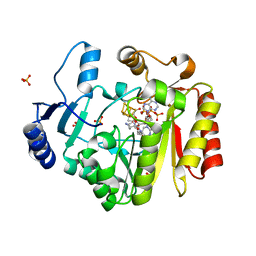 | | Spore photoproduct lyase complexed with dinucleoside spore photoproduct | | Descriptor: | 1-[(2R,4S,5R)-5-(hydroxymethyl)-4-oxidanyl-oxolan-2-yl]-5-[[(5R)-1-[(2R,4S,5R)-5-(hydroxymethyl)-4-oxidanyl-oxolan-2-yl]-5-methyl-2,4-bis(oxidanylidene)-1,3-diazinan-5-yl]methyl]pyrimidine-2,4-dione, IRON/SULFUR CLUSTER, PYROPHOSPHATE 2-, ... | | Authors: | Benjdia, A, Heil, K, Barends, T.R.M, Carell, T, Schlichting, I. | | Deposit date: | 2012-06-06 | | Release date: | 2012-07-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into recognition and repair of UV-DNA damage by Spore Photoproduct Lyase, a radical SAM enzyme.
Nucleic Acids Res., 40, 2012
|
|
4FHG
 
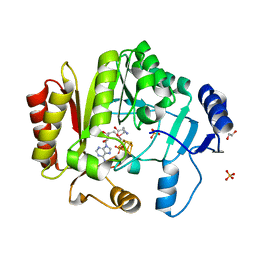 | | Spore photoproduct lyase C140S mutant | | Descriptor: | 1,2-ETHANEDIOL, IRON/SULFUR CLUSTER, SULFATE ION, ... | | Authors: | Benjdia, A, Heil, K, Barends, T.R.M, Carell, T, Schlichting, I. | | Deposit date: | 2012-06-06 | | Release date: | 2012-07-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into recognition and repair of UV-DNA damage by Spore Photoproduct Lyase, a radical SAM enzyme.
Nucleic Acids Res., 40, 2012
|
|
7LYE
 
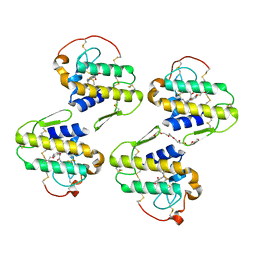 | |
4FGI
 
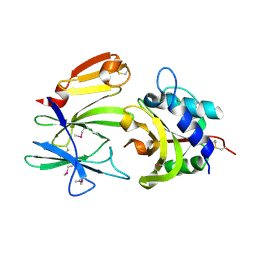 | | Structure of the effector - immunity system Tse1 / Tsi1 from Pseudomonas aeruginosa | | Descriptor: | Tse1, Tsi1 | | Authors: | Benz, J, Sendlmeier, C, Barends, T.R.M, Meinhart, A. | | Deposit date: | 2012-06-04 | | Release date: | 2012-06-20 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Insights into the Effector - Immunity System Tse1/Tsi1 from Pseudomonas aeruginosa.
Plos One, 7, 2012
|
|
4FHF
 
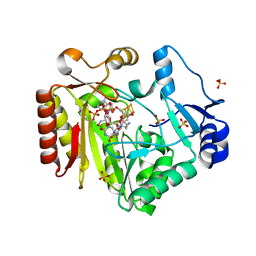 | | Spore photoproduct lyase C140A mutant with dinucleoside spore photoproduct | | Descriptor: | 1-[(2R,4S,5R)-5-(hydroxymethyl)-4-oxidanyl-oxolan-2-yl]-5-[[(5R)-1-[(2R,4S,5R)-5-(hydroxymethyl)-4-oxidanyl-oxolan-2-yl]-5-methyl-2,4-bis(oxidanylidene)-1,3-diazinan-5-yl]methyl]pyrimidine-2,4-dione, IRON/SULFUR CLUSTER, PYROPHOSPHATE 2-, ... | | Authors: | Benjdia, A, Heil, K, Barends, T.R.M, Carell, T, Schlichting, I. | | Deposit date: | 2012-06-06 | | Release date: | 2012-07-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into recognition and repair of UV-DNA damage by Spore Photoproduct Lyase, a radical SAM enzyme.
Nucleic Acids Res., 40, 2012
|
|
7M02
 
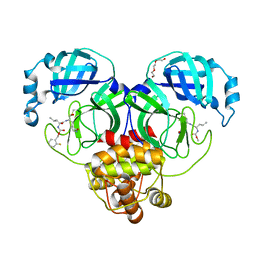 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 17c | | Descriptor: | (1R,2S)-2-((S)-2-((((2-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((2-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7LZV
 
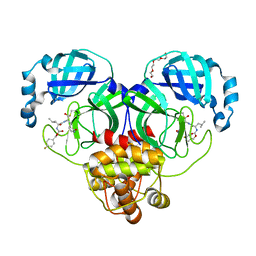 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 19b | | Descriptor: | (1R,2S)-2-((S)-2-((((4-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((4-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7M00
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 13c | | Descriptor: | (1R,2S)-2-((S)-2-((((2-(4,4-difluorocyclohexyl)propan-2-yl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((2-(4,4-difluorocyclohexyl)propan-2-yl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl)propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7LZW
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 20b (deuterated analog of 19b) | | Descriptor: | (1R,2S)-2-((S)-2-((((4-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((4-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
