4PN9
 
 | | A de novo designed hexameric coiled coil CC-Hex2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CC-Hex2 | | Authors: | Wood, C.W, Burton, A.J, Thomson, A.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2014-05-23 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Computational design of water-soluble alpha-helical barrels.
Science, 346, 2014
|
|
4PNA
 
 | | A de novo designed heptameric coiled coil CC-Hept | | Descriptor: | CC-Hept, GLYCEROL | | Authors: | Burton, A.J, Wood, C.W, Thomson, A.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2014-05-23 | | Release date: | 2014-10-22 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Computational design of water-soluble alpha-helical barrels.
Science, 346, 2014
|
|
3IM2
 
 | |
3NN8
 
 | |
2FT1
 
 | | Bacteriophage HK97 Head II | | Descriptor: | major capsid protein | | Authors: | Gan, L, Speir, J.A, Conway, J.F, Lander, G, Cheng, N, Firek, B.A, Hendrix, R.W, Duda, R.L, Liljas, L, Johnson, J.E. | | Deposit date: | 2006-01-23 | | Release date: | 2006-02-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Capsid Conformational Sampling in HK97 Maturation Visualized by X-Ray Crystallography and Cryo-EM.
Structure, 14, 2006
|
|
4P7B
 
 | | Crystal structure of S. typhimurium peptidyl-tRNA hydrolase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Peptidyl-tRNA hydrolase | | Authors: | Vandavasi, V.G, McFeeters, R.L, Taylor-Creel, K, Coates, L, McFeeters, H. | | Deposit date: | 2014-03-26 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recombinant production, crystallization and X-ray crystallographic structure determination of peptidyl-tRNA hydrolase from Salmonella typhimurium.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3NZQ
 
 | | Crystal Structure of Biosynthetic arginine decarboxylase ADC (SpeA) from Escherichia coli, Northeast Structural Genomics Consortium Target ER600 | | Descriptor: | Biosynthetic arginine decarboxylase, SULFATE ION | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-07-16 | | Release date: | 2010-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of bacterial biosynthetic arginine decarboxylases.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2FS3
 
 | | Bacteriophage HK97 K169Y Head I | | Descriptor: | Major capsid protein | | Authors: | Gan, L, Speir, J.A, Conway, J.F, Lander, G, Cheng, N, Firek, B.A, Hendrix, R.W, Duda, R.L, Liljas, L, Johnson, J.E. | | Deposit date: | 2006-01-20 | | Release date: | 2006-02-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Capsid Conformational Sampling in HK97 Maturation Visualized by X-Ray Crystallography and Cryo-EM.
Structure, 14, 2006
|
|
2FVD
 
 | | Cyclin Dependent Kinase 2 (CDK2) with diaminopyrimidine inhibitor | | Descriptor: | (4-AMINO-2-{[1-(METHYLSULFONYL)PIPERIDIN-4-YL]AMINO}PYRIMIDIN-5-YL)(2,3-DIFLUORO-6-METHOXYPHENYL)METHANONE, Cell division protein kinase 2 | | Authors: | Crowther, R.L, Lukacs, C.M, Kammlott, R.U. | | Deposit date: | 2006-01-30 | | Release date: | 2006-10-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of [4-Amino-2-(1-methanesulfonylpiperidin-4-ylamino)pyrimidin-5-yl](2,3-difluoro-6- methoxyphenyl)methanone (R547), a potent and selective cyclin-dependent kinase inhibitor with significant in vivo antitumor activity.
J.Med.Chem., 49, 2006
|
|
3IT3
 
 | | Crystal Structure Francisella tularensis histidine acid phosphatase D261A mutant complexed with substrate 3'-AMP | | Descriptor: | Acid phosphatase, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl] dihydrogen phosphate | | Authors: | Singh, H, Felts, R.L, Reilly, T.J, Tanner, J.J. | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of the histidine acid phosphatase from Francisella tularensis provide insight into substrate recognition.
J.Mol.Biol., 394, 2009
|
|
4PNB
 
 | | A de novo designed hexameric coiled coil CC-Hex3. | | Descriptor: | CC-Hex3 | | Authors: | Wood, C.W, Burton, A.J, Thomson, A.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2014-05-23 | | Release date: | 2014-10-22 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Computational design of water-soluble alpha-helical barrels.
Science, 346, 2014
|
|
2GG0
 
 | | Novel bacterial methionine aminopeptidase inhibitors | | Descriptor: | COBALT (II) ION, METHYL N-{(2S,3R)-3-AMINO-2-HYDROXY-3-[4-(TRIFLUOROMETHYL)PHENYL]PROPANOYL}ALANYLGLYCINATE, Methionine aminopeptidase, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Serendipitous discovery of novel bacterial methionine aminopeptidase inhibitors.
Proteins, 66, 2007
|
|
2GGB
 
 | | Novel bacterial methionine aminopeptidase inhibitors | | Descriptor: | COBALT (II) ION, METHYL N-[(2S,3R)-3-AMINO-2-HYDROXYHEPTANOYL]-L-SERYL-L-LEUCINATE, Methionine aminopeptidase, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Serendipitous discovery of novel bacterial methionine aminopeptidase inhibitors.
Proteins, 66, 2007
|
|
2GG2
 
 | | Novel bacterial methionine aminopeptidase inhibitors | | Descriptor: | 5-IMINO-4-(3-TRIFLUOROMETHYL-PHENYLAZO)-5H-PYRAZOL-3-YLAMINE, COBALT (II) ION, Methionine aminopeptidase, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Serendipitous discovery of novel bacterial methionine aminopeptidase inhibitors.
Proteins, 66, 2007
|
|
2FTE
 
 | | Bacteriophage HK97 Expansion Intermediate IV | | Descriptor: | major capsid protein | | Authors: | Gan, L, Speir, J.A, Conway, J.F, Lander, G, Cheng, N, Firek, B.A, Hendrix, R.W, Duda, R.L, Liljas, L, Johnson, J.E. | | Deposit date: | 2006-01-24 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY | | Cite: | Capsid Conformational Sampling in HK97 Maturation Visualized by X-Ray Crystallography and Cryo-EM.
Structure, 14, 2006
|
|
6UFL
 
 | | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199Q mutant) from Amycolatopsis mediterranei (AmGH128_I) in the complex with laminarihexaose | | Descriptor: | Glyco_hydro_cc domain-containing protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Cordeiro, R.L, Domingues, M.N, Vieira, P.S, Santos, C.R, Murakami, M.T. | | Deposit date: | 2019-09-24 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
2G8E
 
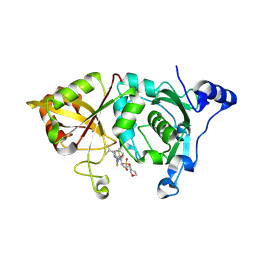 | | Calpain 1 proteolytic core in complex with SNJ-1715, a cyclic hemiacetal-type inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Calpain-1 catalytic subunit, ... | | Authors: | Cuerrier, D, Moldoveanu, T, Davies, P.L, Campbell, R.L. | | Deposit date: | 2006-03-02 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Calpain Inhibition by alpha-Ketoamide and Cyclic Hemiacetal Inhibitors Revealed by X-ray Crystallography
Biochemistry, 45, 2006
|
|
6UFZ
 
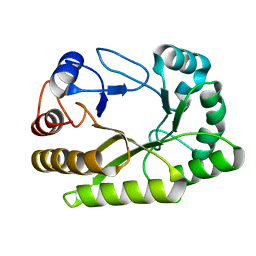 | | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199Q mutant) from Amycolatopsis mediterranei (AmGH128_I) | | Descriptor: | Glyco_hydro_cc domain-containing protein | | Authors: | Cordeiro, R.L, Domingues, M.N, Vieira, P.S, Santos, C.R, Murakami, M.T. | | Deposit date: | 2019-09-25 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
2GG7
 
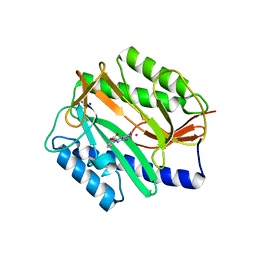 | | Novel bacterial methionine aminopeptidase inhibitors | | Descriptor: | 3-(5-AMINO-3-IMINO-3H-PYRAZOL-4-YLAZO)-BENZOIC ACID, COBALT (II) ION, Methionine aminopeptidase, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Serendipitous discovery of novel bacterial methionine aminopeptidase inhibitors.
Proteins, 66, 2007
|
|
6UNA
 
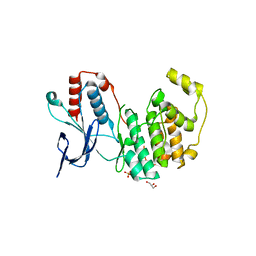 | | Crystal structure of inactive p38gamma | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 12, SULFATE ION | | Authors: | Aoto, P.C, Stanfield, R.L, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2019-10-11 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | A Dynamic Switch in Inactive p38 gamma Leads to an Excited State on the Pathway to an Active Kinase.
Biochemistry, 58, 2019
|
|
6UBA
 
 | | Crystal structure of a GH128 (subgroup VI) exo-beta-1,3-glucanase from Aureobasidium namibiae (AnGH128_VI) in complex with laminaritriose | | Descriptor: | Glyco_hydro_cc domain-containing protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Santos, C.R, Vieira, P.S, Domingues, M.N, Cordeiro, R.L, Tomazini, A, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
2G3P
 
 | |
2GG8
 
 | | Novel bacterial methionine aminopeptidase inhibitors | | Descriptor: | COBALT (II) ION, METHYL N-[(2S,3R)-3-AMINO-2-HYDROXY-3-(4-METHYLPHENYL)PROPANOYL]-D-ALANYL-D-LEUCINATE, Methionine aminopeptidase, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Serendipitous discovery of novel bacterial methionine aminopeptidase inhibitors.
Proteins, 66, 2007
|
|
2GHL
 
 | | Mutant Mus Musculus P38 Kinase Domain in Complex with Inhibitor PG-874743 | | Descriptor: | 3-(2-CHLOROPHENYL)-1-(2-{[(1S)-2-HYDROXY-1,2-DIMETHYLPROPYL]AMINO}PYRIMIDIN-4-YL)-1-(4-METHOXYPHENYL)UREA, Mitogen-activated protein kinase 14 | | Authors: | Walter, R.L, Mekel, M.J, Evdokimov, A.G, Pokross, M.E, Brugel, T.A. | | Deposit date: | 2006-03-27 | | Release date: | 2006-04-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Development of N-2,4-pyrimidine-N-phenyl-N'-phenyl ureas as inhibitors of tumor necrosis factor alpha (TNF-alpha) synthesis. Part 1.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5KBM
 
 | | Candida Albicans Superoxide Dismutase 5 (SOD5), D113N Mutant | | Descriptor: | COPPER (I) ION, Cell surface Cu-only superoxide dismutase 5, SULFATE ION | | Authors: | Galaleldeen, A, Peterson, R.L, Villarreal, J, Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-06-03 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.416 Å) | | Cite: | The Phylogeny and Active Site Design of Eukaryotic Copper-only Superoxide Dismutases.
J.Biol.Chem., 291, 2016
|
|
