4H8G
 
 | |
4HN1
 
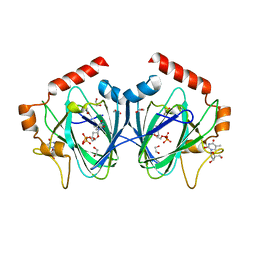 | | Crystal Structure of H60N/Y130F double mutant of ChmJ, a 3'-monoepimerase from Streptomyces bikiniensis in complex with dTDP | | Descriptor: | 1,2-ETHANEDIOL, Putative 3-epimerase in D-allose pathway, THYMIDINE, ... | | Authors: | Holden, H.M, Kubiak, R.L. | | Deposit date: | 2012-10-18 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Studies on a 3'-Epimerase Involved in the Biosynthesis of dTDP-6-deoxy-d-allose.
Biochemistry, 51, 2012
|
|
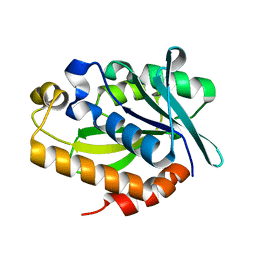
4FYJ
 
 | |
4GC7
 
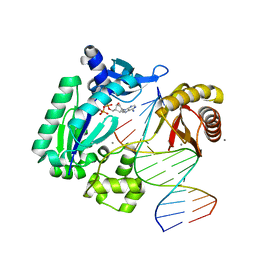 | | Crystal structure of Dpo4 in complex with S-MC-dADP opposite dT | | Descriptor: | CALCIUM ION, DNA (5'-D(*G*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*CP*C)-3'), DNA (5'-D(*TP*CP*AP*TP*GP*GP*AP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Eoff, R.L, Ketkar, A, Banerjee, S, Zafar, M.K. | | Deposit date: | 2012-07-29 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Differential furanose selection in the active sites of archaeal DNA polymerases probed by fixed-conformation nucleotide analogues.
Biochemistry, 51, 2012
|
|
4FXD
 
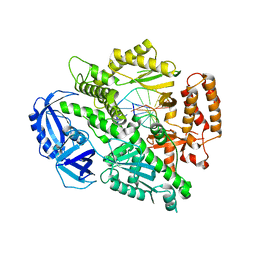 | | Crystal structure of yeast DNA polymerase alpha bound to DNA/RNA | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*CP*GP*CP*TP*GP*CP*CP*CP*GP*CP*CP*T)-3'), DNA polymerase alpha catalytic subunit A, RNA (5'-R(*AP*GP*GP*CP*GP*GP*GP*CP*AP*G)-3') | | Authors: | Perera, R.L, Torella, R, Klinge, S, Kilkenny, M.L, Maman, J.D, Pellegrini, L. | | Deposit date: | 2012-07-03 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanism for priming DNA synthesis by yeast DNA Polymerase alpha
eLife, 2, 2013
|
|
6HAL
 
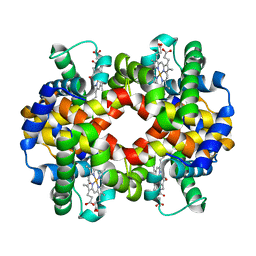 | | Human carbonmonoxy hemoglobin SFX dataset | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Doak, B, Gorel, A, Foucar, L, Barends, T.R.M, Gruenbein, M.L, Hilpert, M, Kloos, M, Nass Kovacs, G, Roome, C.M, Shoeman, R.L, Stricker, M, Tono, K, You, D, Ueda, K, Sherrell, D.A, Owen, R.L, Schlichting, I. | | Deposit date: | 2018-08-07 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallography on a chip - without the chip: sheet-on-sheet sandwich.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5SXY
 
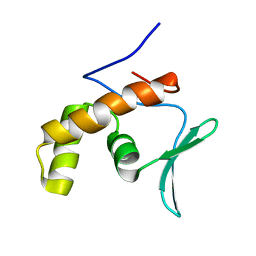 | |
4W85
 
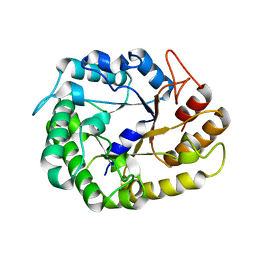 | | Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from ruminal metagenomic library, in complex with glucose | | Descriptor: | MAGNESIUM ION, Xyloglucan-specific endo-beta-1,4-glucanase, beta-D-glucopyranose | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
6E12
 
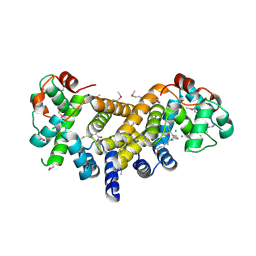 | | Crystal Structure of the Alr8543 protein in complex with Oleic Acid and magnesium ion from Nostoc sp. PCC 7120, Northeast Structural Genomics Consortium Target NsR141 | | Descriptor: | Alr8543 protein, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Xiao, R, Ciccosanti, C, Patel, D, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-07-09 | | Release date: | 2018-07-25 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Alr8543 protein in complex with Oleic Acid and magnesium ion from Nostoc sp. PCC 7120, Northeast Structural Genomics Consortium Target NsR141
To Be Published
|
|
3JT0
 
 | | Crystal Structure of the C-terminal fragment (426-558) Lamin-B1 from Homo sapiens, Northeast Structural Genomics Consortium Target HR5546A | | Descriptor: | Lamin-B1 | | Authors: | Kuzin, A, Abashidze, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-11 | | Release date: | 2009-09-22 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Northeast Structural Genomics Consortium Target HR5546A
To be Published
|
|
8HVP
 
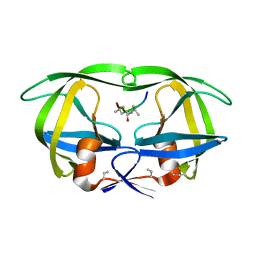 | | STRUCTURE AT 2.5-ANGSTROMS RESOLUTION OF CHEMICALLY SYNTHESIZED HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 PROTEASE COMPLEXED WITH A HYDROXYETHYLENE*-BASED INHIBITOR | | Descriptor: | HIV-1 PROTEASE, INHIBITOR VAL-SER-GLN-ASN-LEU-PSI(CH(OH)-CH2)-VAL-ILE-VAL (U-85548E) | | Authors: | Jaskolski, M, Miller, M, Tomasselli, A.G, Sawyer, T.K, Staples, D.G, Heinrikson, R.L, Schneider, J, Kent, S.B.H, Wlodawer, A. | | Deposit date: | 1990-10-26 | | Release date: | 1993-10-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure at 2.5-A resolution of chemically synthesized human immunodeficiency virus type 1 protease complexed with a hydroxyethylene-based inhibitor.
Biochemistry, 30, 1991
|
|
8C78
 
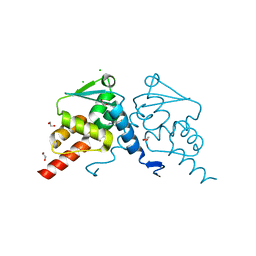 | | Crystal structure of human BCL6 BTB domain in complex with compound CCT374705 | | Descriptor: | (2~{S})-10-[(3-chloranyl-2-fluoranyl-pyridin-4-yl)amino]-2-cyclopropyl-3,3-bis(fluoranyl)-7-methyl-2,4-dihydro-1~{H}-[1,4]oxazepino[2,3-c]quinolin-6-one, 1,2-ETHANEDIOL, B-cell lymphoma 6 protein, ... | | Authors: | Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2023-01-13 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of an In Vivo Chemical Probe for BCL6 Inhibition by Optimization of Tricyclic Quinolinones.
J.Med.Chem., 66, 2023
|
|
6XSS
 
 | | CryoEM structure of designed helical fusion protein C4_nat_HFuse-7900 | | Descriptor: | C4_nat_HFuse-7900 | | Authors: | Redler, R.L, Edman, N.I, Baker, D, Ekiert, D, Bhabha, G. | | Deposit date: | 2020-07-16 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
2DOQ
 
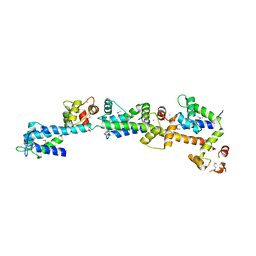 | | crystal structure of Sfi1p/Cdc31p complex | | Descriptor: | CALCIUM ION, Cell division control protein 31, SFI1p | | Authors: | Li, S, Sandercock, A.M, Conduit, P.T, Robinson, C.V, Williams, R.L, Kilmartin, J.V. | | Deposit date: | 2006-05-03 | | Release date: | 2006-06-27 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication.
J.Cell Biol., 173, 2006
|
|
5OQ4
 
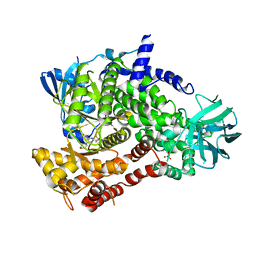 | | PQR309 - a Potent, Brain-Penetrant, Orally Bioavailable, pan-Class I PI3K/mTOR Inhibitor as Clinical Candidate in Oncology | | Descriptor: | 5-(4,6-dimorpholin-4-yl-1,3,5-triazin-2-yl)-4-(trifluoromethyl)pyridin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Williams, R.L, Zhang, X. | | Deposit date: | 2017-08-10 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 5-(4,6-Dimorpholino-1,3,5-triazin-2-yl)-4-(trifluoromethyl)pyridin-2-amine (PQR309), a Potent, Brain-Penetrant, Orally Bioavailable, Pan-Class I PI3K/mTOR Inhibitor as Clinical Candidate in Oncology.
J. Med. Chem., 60, 2017
|
|
5T9W
 
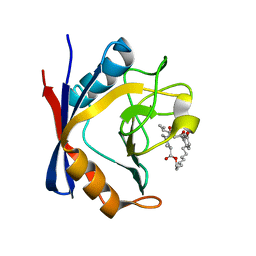 | | Discovery of a Potent Cyclophilin Inhibitor (Compound 5) based on Structural Simplification of Sanglifehrin A | | Descriptor: | 3-[(3-hydroxyphenyl)methyl]-6-(propan-2-yl)-19-oxa-1,4,7,25-tetraazabicyclo[19.3.1]pentacosa-13,15-diene-2,5,8,20-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Steadman, V, Pettit, P, Schmitz, U, Mackman, R.L, Schultz, B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Potent Cyclophilin Inhibitors Based on the Structural Simplification of Sanglifehrin A.
J. Med. Chem., 60, 2017
|
|
5T9U
 
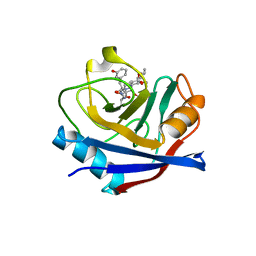 | | Discovery of a Potent Cyclophilin Inhibitor (Compound 3) based on Structural Simplification of Sanglifehrin A | | Descriptor: | 3-[(3-hydroxyphenyl)methyl]-10,12-dimethoxy-9,11-dimethyl-6-(propan-2-yl)-19-oxa-1,4,7,25-tetraazabicyclo[19.3.1]pentacosa-13,15-diene-2,5,8,20-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Steadman, V, Pettit, S, Schmitz, U, Mackman, R.L, Schultz, B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Discovery of Potent Cyclophilin Inhibitors Based on the Structural Simplification of Sanglifehrin A.
J. Med. Chem., 60, 2017
|
|
5T9Z
 
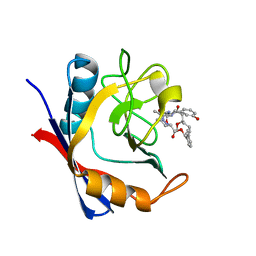 | | Discovery of a Potent Cyclophilin Inhibitor (Compound 6) based on Structural Simplification of Sanglifehrin A | | Descriptor: | 11-[(3-hydroxyphenyl)methyl]-18-methoxy-17-methyl-14-(propan-2-yl)-3-oxa-9,12,15,28-tetraazatricyclo[21.3.1.1~5,9~]octacosa-1(27),21,23,25-tetraene-4,10,13,16-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Steadman, V, Pettit, S, Schmitz, U, Mackman, R.L, Schultz, B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of Potent Cyclophilin Inhibitors Based on the Structural Simplification of Sanglifehrin A.
J. Med. Chem., 60, 2017
|
|
5T3S
 
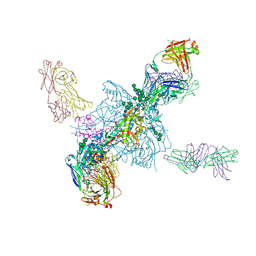 | | HIV gp140 trimer MD39-10MUTA in complex with Fabs PGT124 and 35022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2016-08-26 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | HIV Vaccine Design to Target Germline Precursors of Glycan-Dependent Broadly Neutralizing Antibodies.
Immunity, 45, 2016
|
|
4N6U
 
 | | Adhiron: a stable and versatile peptide display scaffold - truncated adhiron | | Descriptor: | Adhiron | | Authors: | Mcpherson, M, Tomlinson, D, Owen, R.L, Nettleship, J.E, Owens, R.J. | | Deposit date: | 2013-10-14 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Adhiron: a stable and versatile peptide display scaffold for molecular recognition applications.
Protein Eng.Des.Sel., 27, 2014
|
|
4N9U
 
 | | The role of lysine 200 in the human farnesyl pyrophosphate synthase catalytic mechanism and the mode of inhibition by the nitrogen-containing bisphosphonates | | Descriptor: | 1,2-ETHANEDIOL, 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, Farnesyl pyrophosphate synthase, ... | | Authors: | Tsoumpra, M.K, Muniz, J.R.C, Barnett, B.L, Pilka, E, Kwaasi, A.A, Kavanagh, K.L, Evdokimov, A, Walter, R.L, Ebetino, F.H, Oppermann, U, Russell, R.G.G, Dunford, J.E. | | Deposit date: | 2013-10-21 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The role of lysine 200 in the human farnesyl pyrophosphate synthase catalytic mechanism and the mode of inhibition by the nitrogen-containing bisphosphonates
TO BE PUBLISHED
|
|
6M87
 
 | | Fab 10A6 in complex with MPTS | | Descriptor: | 8-methoxypyrene-1,3,6-trisulfonic acid, Fab 10A6 heavy chain, Fab 10A6 light chain, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-08-21 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Structure and Dynamics of Stacking Interactions in an Antibody Binding Site.
Biochemistry, 58, 2019
|
|
6M94
 
 | | Monophosphorylated pSer33 b-Catenin peptide bound to b-TrCP/Skp1 Complex | | Descriptor: | Catenin beta-1, DIMETHYL SULFOXIDE, F-box/WD repeat-containing protein 1A, ... | | Authors: | Simonetta, K.R, Clifton, M.C, Walter, R.L, Ranieri, G.M, Carter, J.J. | | Deposit date: | 2018-08-22 | | Release date: | 2019-04-03 | | Last modified: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Prospective discovery of small molecule enhancers of an E3 ligase-substrate interaction.
Nat Commun, 10, 2019
|
|
7QZG
 
 | | SFX structure of dye-type peroxidase DtpB N245A variant in the ferric state | | Descriptor: | Dyp-type peroxidase family, MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lucic, M, Worrall, J.A.R, Hough, M.A, Shilova, A, Axford, D.A, Owen, R.L, Tosha, T, Sugimoto, H, Owada, S. | | Deposit date: | 2022-01-31 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial Femtosecond Crystallography Reveals the Role of Water in the One- or Two-Electron Redox Chemistry of Compound I in the Catalytic Cycle of the B-Type Dye-Decolorizing Peroxidase DtpB.
Acs Catalysis, 12, 2022
|
|
4W8A
 
 | | Crystal structure of XEG5B, a GH5 xyloglucan-specific beta-1,4-glucanase from ruminal metagenomic library, in the native form | | Descriptor: | Exo-xyloglucanase, GLYCEROL, SULFATE ION | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
