1BV1
 
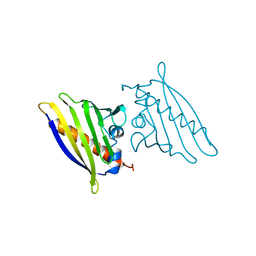 | | BIRCH POLLEN ALLERGEN BET V 1 | | Descriptor: | BET V 1 | | Authors: | Gajhede, M, Osmark, P, Poulsen, F.M, Ipsen, H, Larson, J.N, Joostvan, R.J, Schou, C, Lowenstein, H, Spangfort, M.D. | | Deposit date: | 1997-07-08 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray and NMR structure of Bet v 1, the origin of birch pollen allergy.
Nat.Struct.Biol., 3, 1996
|
|
2WJW
 
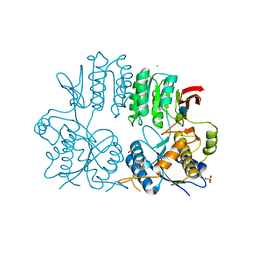 | | Crystal structure of the human ionotropic glutamate receptor GluR2 ATD region at 1.8 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CHLORIDE ION, ... | | Authors: | Clayton, A, Siebold, C, Gilbert, R.J.C, Sutton, G.C, Harlos, K, McIlhinney, R.A.J, Jones, E.Y, Aricescu, A.R. | | Deposit date: | 2009-06-01 | | Release date: | 2009-08-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Glur2 Amino-Terminal Domain Provides Insights Into the Architecture and Assembly of Ionotropic Glutamate Receptors.
J.Mol.Biol., 392, 2009
|
|
1IS9
 
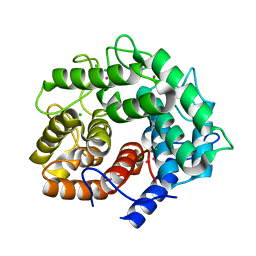 | | Endoglucanase A from Clostridium thermocellum at atomic resolution | | Descriptor: | CHLORIDE ION, MERCURY (II) ION, endoglucanase A | | Authors: | Schmidt, A, Gonzalez, A, Morris, R.J, Costabel, M, Alzari, P.M, Lamzin, V.S. | | Deposit date: | 2001-11-26 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Advantages of high-resolution phasing: MAD to atomic resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1F29
 
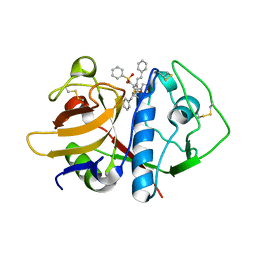 | | CRYSTAL STRUCTURE ANALYSIS OF CRUZAIN BOUND TO A VINYL SULFONE DERIVED INHIBITOR (I) | | Descriptor: | 3-[[N-[MORPHOLIN-N-YL]-CARBONYL]-PHENYLALANINYL-AMINO]-5- PHENYL-PENTANE-1-SULFONYLBENZENE, CRUZAIN | | Authors: | Brinen, L.S, Hansell, E, Roush, W.R, McKerrow, J.H, Fletterick, R.J. | | Deposit date: | 2000-05-23 | | Release date: | 2000-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A target within the target: probing cruzain's P1' site to define structural determinants for the Chagas' disease protease.
Structure Fold.Des., 8, 2000
|
|
1L5R
 
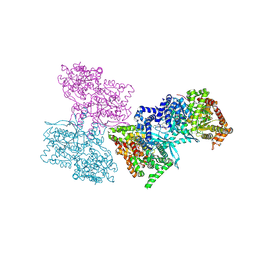 | | Human liver glycogen phosphorylase a complexed with riboflavin, N-Acetyl-beta-D-Glucopyranosylamine and CP-403,700 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, N-acetyl-beta-D-glucopyranosylamine, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Ekstrom, J.L, Pauly, T.A, Carty, M.D, Soeller, W.C, Culp, J, Danley, D.E, Hoover, D.J, Treadway, J.L, Gibbs, E.M, Fletterick, R.J, Day, Y.S.N, Myszka, D.G, Rath, V.L. | | Deposit date: | 2002-03-07 | | Release date: | 2002-12-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-activity analysis of the purine binding
site of human liver glycogen phosphorylase.
Chem.Biol., 9, 2002
|
|
1G9Y
 
 | | HOMING ENDONUCLEASE I-CREI / DNA SUBSTRATE COMPLEX WITH CALCIUM | | Descriptor: | 5'-D(*CP*GP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*TP*TP*GP*C)-3', 5'-D(*GP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*TP*CP*G)-3', CALCIUM ION, ... | | Authors: | Chevalier, B, Monnat, R.J, Stoddard, B.L. | | Deposit date: | 2000-11-28 | | Release date: | 2001-04-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The homing endonuclease I-CreI uses three metals, one of which is shared between the two active sites.
Nat.Struct.Biol., 8, 2001
|
|
1BRA
 
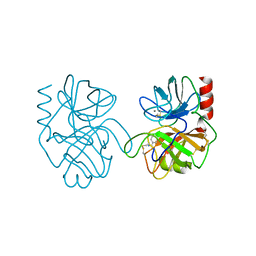 | |
1G9Z
 
 | | LAGLIDADG HOMING ENDONUCLEASE I-CREI / DNA PRODUCT COMPLEX WITH MAGNESIUM | | Descriptor: | 5'-D(*CP*GP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*C)-3', 5'-D(*GP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*A)-3', 5'-D(P*GP*AP*CP*AP*GP*TP*TP*TP*CP*G)-3', ... | | Authors: | Chevalier, B, Monnat, R.J, Stoddard, B.L. | | Deposit date: | 2000-11-28 | | Release date: | 2001-04-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The homing endonuclease I-CreI uses three metals, one of which is shared between the two active sites.
Nat.Struct.Biol., 8, 2001
|
|
7L4U
 
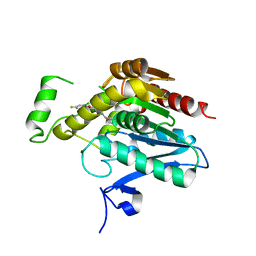 | | Crystal structure of human monoacylglycerol lipase in complex with compound 1h | | Descriptor: | (5S)-5-(3-{4-[(2-chloro-4-fluorophenoxy)methyl]piperidin-1-yl}-3-oxopropyl)pyrrolidin-2-one, CHLORIDE ION, Monoglyceride lipase | | Authors: | Qin, L, Lane, W, Skene, R.J, Dougan, D. | | Deposit date: | 2020-12-21 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design and Synthesis of Novel Spiro Derivatives as Potent and Reversible Monoacylglycerol Lipase (MAGL) Inhibitors: Bioisosteric Transformation from 3-Oxo-3,4-dihydro-2 H -benzo[ b ][1,4]oxazin-6-yl Moiety.
J.Med.Chem., 64, 2021
|
|
7L4W
 
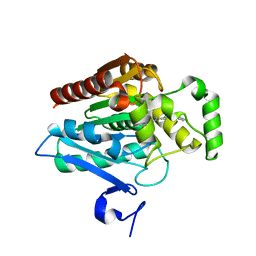 | | Crystal structure of human monoacylglycerol lipase in complex with compound 2d | | Descriptor: | (2s,4R)-2-{4-[(2-chloro-4-fluorophenoxy)methyl]piperidine-1-carbonyl}-7-oxa-5-azaspiro[3.4]octan-6-one, Monoglyceride lipase | | Authors: | Qin, L, Gay, S.C, Lane, W, Skene, R.J. | | Deposit date: | 2020-12-21 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and Synthesis of Novel Spiro Derivatives as Potent and Reversible Monoacylglycerol Lipase (MAGL) Inhibitors: Bioisosteric Transformation from 3-Oxo-3,4-dihydro-2 H -benzo[ b ][1,4]oxazin-6-yl Moiety.
J.Med.Chem., 64, 2021
|
|
7L4T
 
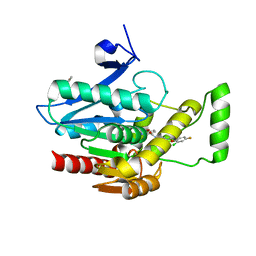 | | Crystal structure of human monoacylglycerol lipase in complex with compound 1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-{4-[(2-chloro-4-fluorophenoxy)methyl]piperidine-1-carbonyl}-2H-1,4-benzoxazin-3(4H)-one, ACETATE ION, ... | | Authors: | Qin, L, Gay, S.C, Lane, W, Skene, R.J. | | Deposit date: | 2020-12-21 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and Synthesis of Novel Spiro Derivatives as Potent and Reversible Monoacylglycerol Lipase (MAGL) Inhibitors: Bioisosteric Transformation from 3-Oxo-3,4-dihydro-2 H -benzo[ b ][1,4]oxazin-6-yl Moiety.
J.Med.Chem., 64, 2021
|
|
7L50
 
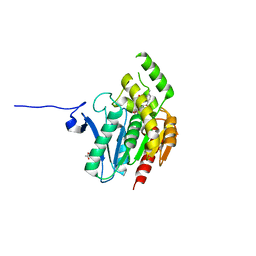 | | Crystal structure of human monoacylglycerol lipase in complex with compound 4f | | Descriptor: | (2s,4R)-2-{3-[(3-chloro-4-methylphenyl)methoxy]azetidine-1-carbonyl}-7-oxa-5-azaspiro[3.4]octan-6-one, ACETATE ION, Monoglyceride lipase | | Authors: | Qin, L, Lane, W, Skene, R.J. | | Deposit date: | 2020-12-21 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Synthesis of Novel Spiro Derivatives as Potent and Reversible Monoacylglycerol Lipase (MAGL) Inhibitors: Bioisosteric Transformation from 3-Oxo-3,4-dihydro-2 H -benzo[ b ][1,4]oxazin-6-yl Moiety.
J.Med.Chem., 64, 2021
|
|
8PJN
 
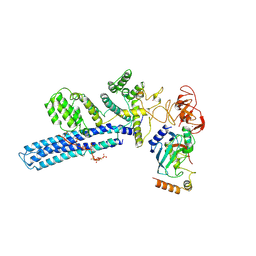 | | Catalytic module of human CTLH E3 ligase bound to multiphosphorylated UBE2H~ubiquitin | | Descriptor: | E3 ubiquitin-protein transferase MAEA, E3 ubiquitin-protein transferase RMND5A, Ubiquitin, ... | | Authors: | Chrustowicz, J, Sherpa, D, Prabu, R.J, Schulman, B.A. | | Deposit date: | 2023-06-23 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Multisite phosphorylation dictates selective E2-E3 pairing as revealed by Ubc8/UBE2H-GID/CTLH assemblies.
Mol.Cell, 84, 2024
|
|
8PMQ
 
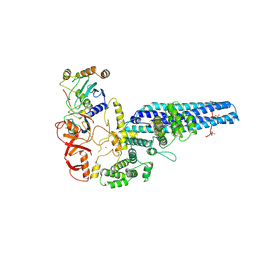 | | Catalytic module of yeast GID E3 ligase bound to multiphosphorylated Ubc8~ubiquitin | | Descriptor: | E3 ubiquitin-protein ligase RMD5, Protein FYV10, Ubiquitin, ... | | Authors: | Chrustowicz, J, Sherpa, D, Prabu, R.J, Schulman, B.A. | | Deposit date: | 2023-06-29 | | Release date: | 2024-01-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Multisite phosphorylation dictates selective E2-E3 pairing as revealed by Ubc8/UBE2H-GID/CTLH assemblies.
Mol.Cell, 84, 2024
|
|
7KVZ
 
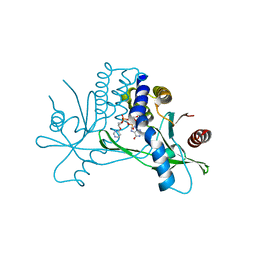 | | Structure of hSTING in complex with novel carbocyclic pyrimidine CDN-2 | | Descriptor: | (2R,5R,7R,8R,10S,12aR,14R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-2,10,16-trihydroxy-14-[(pyrimidin-4-yl)oxy]decahydro-2H,10H-5,8-methano-2lambda~5~,10lambda~5~-cyclopenta[l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Skene, R.J. | | Deposit date: | 2020-11-29 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of Novel Carbocyclic Pyrimidine Cyclic Dinucleotide STING Agonists for Antitumor Immunotherapy Using Systemic Intravenous Route.
J.Med.Chem., 64, 2021
|
|
8QBB
 
 | | Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture +PIP2/-sterol (DOPC, DOPE, DOPS, PI(4,5)P2 50:20:20:10) | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Sphingolipid long chain base-responsive protein PIL1 | | Authors: | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Cryo-EM architecture of a near-native stretch-sensitive membrane microdomain.
Nature, 2024
|
|
8QB9
 
 | | Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture -PIP2/+sterol (DOPC, DOPE, DOPS, cholesterol 30:20:20:30) | | Descriptor: | Sphingolipid long chain base-responsive protein PIL1 | | Authors: | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM architecture of a near-native stretch-sensitive membrane microdomain.
Nature, 2024
|
|
8ATG
 
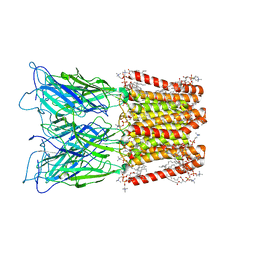 | | Pentameric ligand-gated ion channel GLIC with bound lipids | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Proton-gated ion channel | | Authors: | Bergh, C, Rovsnik, U, Howard, R.J, Lindahl, E. | | Deposit date: | 2022-08-23 | | Release date: | 2023-09-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Discovery of lipid binding sites in a ligand-gated ion channel by integrating simulations and cryo-EM.
Elife, 12, 2024
|
|
8SNI
 
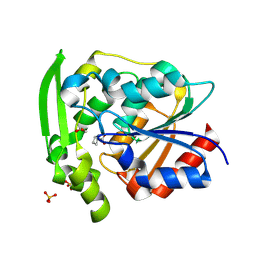 | | Hydroxynitrile Lyase from Hevea brasiliensis with Forty Mutations | | Descriptor: | (S)-hydroxynitrile lyase, 1,2-ETHANEDIOL, BENZOIC ACID, ... | | Authors: | Walsh, M.E, Greenberg, L.R, Kazlauskas, R.J, Pierce, C.T, Aihara, H, Evans, R.L, Shi, K. | | Deposit date: | 2023-04-27 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | To be published
To Be Published
|
|
7NB9
 
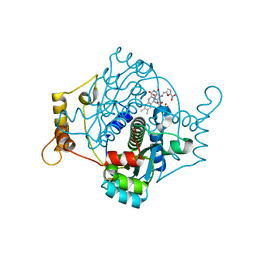 | | E. coli NfsA with nitrofurantoin | | Descriptor: | 1-[(~{E})-(5-nitrofuran-2-yl)methylideneamino]imidazolidine-2,4-dione, DIMETHYL SULFOXIDE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Day, M.D, Jarrom, D, Grainger, A.I, Parr, R.J, Hyde, E.I, White, S.A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN.
Biochem.J., 478, 2021
|
|
7NMP
 
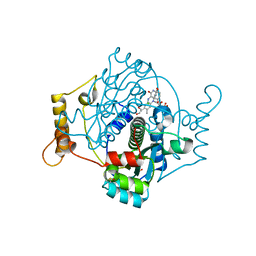 | | E. coli NfsA with hydroquinone | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Day, M.D, Jarrom, D, Parr, R.J, Hyde, E.I, White, S.A. | | Deposit date: | 2021-02-23 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN.
Biochem.J., 478, 2021
|
|
8QFY
 
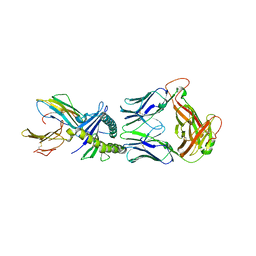 | |
6T6D
 
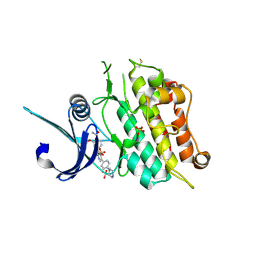 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2149 | | Descriptor: | 2-methoxy-4-[4-methyl-5-(4-piperazin-1-ylphenyl)pyridin-3-yl]benzamide, Activin receptor type I, SULFATE ION | | Authors: | Adamson, R.J, Williams, E.P, Smil, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-10-18 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Targeting ALK2: An Open Science Approach to Developing Therapeutics for the Treatment of Diffuse Intrinsic Pontine Glioma.
J.Med.Chem., 63, 2020
|
|
7NDT
 
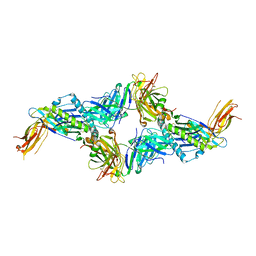 | | UL40:01 TCR in complex with HLA-E with a non-natural amino acid | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, alpha chain E, ... | | Authors: | Pengelly, R.J, Robinson, R.A. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
7NDU
 
 | | Gag:02 TCR in complex with HLA-E featuring a non-natural amino acid | | Descriptor: | Beta-2-microglobulin, Gag6V(276-284 H4C), HLA class I histocompatibility antigen, ... | | Authors: | Pengelly, R.J, Robinson, R.A. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
