3P9Z
 
 | | Crystal structure of uroporphyrinogen-III synthetase from Helicobacter pylori 26695 | | Descriptor: | MALONATE ION, Uroporphyrinogen III cosynthase (HemD) | | Authors: | Nocek, B, Stein, A, Chhor, G, Fenske, R.J, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-10-18 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of uroporphyrinogen-III synthetase from Helicobacter pylori 26695
To be Published
|
|
2L9C
 
 | | Structural insights into the specificity of darcin, an atypical major urinary protein. | | Descriptor: | Darcin | | Authors: | Phelan, M.M, Mclean, L, Beynon, R.J, Hurst, J.L, Lian, L. | | Deposit date: | 2011-02-07 | | Release date: | 2012-02-08 | | Last modified: | 2023-12-06 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the specificity of darcin, an atypical major urinary protein.
To be Published
|
|
8OYT
 
 | | Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '3 up' RBD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H6 nanobody, ... | | Authors: | Weckener, M, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
1A4X
 
 | | PYRR, THE BACILLUS SUBTILIS PYRIMIDINE BIOSYNTHETIC OPERON REPRESSOR, HEXAMERIC FORM | | Descriptor: | PYRIMIDINE OPERON REGULATORY PROTEIN PYRR, SULFATE ION | | Authors: | Tomchick, D.R, Turner, R.J, Switzer, R.W, Smith, J.L. | | Deposit date: | 1998-02-08 | | Release date: | 1998-08-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Adaptation of an enzyme to regulatory function: structure of Bacillus subtilis PyrR, a pyr RNA-binding attenuation protein and uracil phosphoribosyltransferase.
Structure, 6, 1998
|
|
8OPP
 
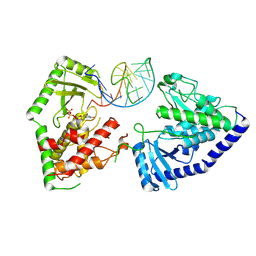 | | Structure of human terminal uridylyltransferase 7 (hTUT7/ZCCHC6) bound with pre-let7g miRNA and UTPalphaS | | Descriptor: | RNA (25-MER), Terminal uridylyltransferase 7, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-sulfanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Yi, G, Ye, M, Gilbert, R.J. | | Deposit date: | 2023-04-07 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs.
Nat.Struct.Mol.Biol., 2024
|
|
8OPS
 
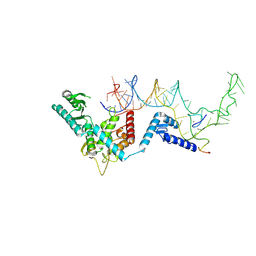 | |
8OST
 
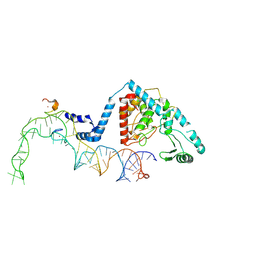 | | Structure of human terminal uridylyltransferase 4 (TUT4, ZCCHC11) in complex with pre-let7g miRNA and Lin28A | | Descriptor: | Protein lin-28 homolog A, Terminal uridylyltransferase 4, ZINC ION, ... | | Authors: | Gilbert, R.J, Yi, G, Ye, M. | | Deposit date: | 2023-04-20 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs.
Nat.Struct.Mol.Biol., 2024
|
|
8OPT
 
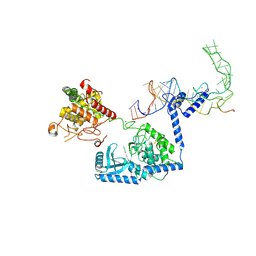 | |
8CYK
 
 | | Crystal structure of hallucinated protein HALC1_878 | | Descriptor: | HALC1_878 | | Authors: | Ragotte, R.J, Bera, A.K, Milles, L.F, Wicky, B.I.M, Baker, D. | | Deposit date: | 2022-05-23 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Robust deep learning-based protein sequence design using ProteinMPNN.
Science, 378, 2022
|
|
8D04
 
 | | Hallucinated C2 protein assembly HALC2_062 | | Descriptor: | HALC2_062 | | Authors: | Ragotte, R.J, Bera, A.K, Wicky, B.I.M, Milles, L.F, Baker, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Hallucinating symmetric protein assemblies.
Science, 378, 2022
|
|
8D07
 
 | | Hallucinated C3 protein assembly HALC3_109 | | Descriptor: | HALC3_109 | | Authors: | Ragotte, R.J, Bera, A.K, Wicky, B.I.M, Milles, L.F, Baker, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Hallucinating symmetric protein assemblies.
Science, 378, 2022
|
|
1ANC
 
 | |
8D05
 
 | | Hallucinated C2 protein assembly HALC2_065 | | Descriptor: | HALC2_065 | | Authors: | Ragotte, R.J, Bera, A.K, Wicky, B.I.M, Milles, L.F, Baker, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Hallucinating symmetric protein assemblies.
Science, 378, 2022
|
|
3OKL
 
 | | Crystal structure of S25-39 in complex with Kdo(2.8)Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, S25-39 Fab (IgG1k) heavy chain, S25-39 Fab (IgG1k) light chain, ... | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2010-08-25 | | Release date: | 2011-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Common NH53K Mutation in the Combining Site of Antibodies Raised against Chlamydial LPS Glycoconjugates Significantly Increases Avidity.
Biochemistry, 50, 2011
|
|
8D08
 
 | | Hallucinated C4 protein assembly HALC4_135 | | Descriptor: | HALC4_135 | | Authors: | Ragotte, R.J, Bera, A.K, Wicky, B.I.M, Milles, L.F, Baker, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Hallucinating symmetric protein assemblies.
Science, 378, 2022
|
|
1A98
 
 | | XPRTASE FROM E. COLI COMPLEXED WITH GMP | | Descriptor: | XANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE | | Authors: | Vos, S, Parry, R.J, Burns, M.R, De Jersey, J, Martin, J.L. | | Deposit date: | 1998-04-16 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of free and complexed forms of Escherichia coli xanthine-guanine phosphoribosyltransferase.
J.Mol.Biol., 282, 1998
|
|
8D03
 
 | | Hallucinated C2 protein assembly HALC2_068 | | Descriptor: | HALC2_068 | | Authors: | Ragotte, R.J, Bera, A.K, Wicky, B.I.M, Milles, L.F, Baker, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hallucinating symmetric protein assemblies.
Science, 378, 2022
|
|
8D09
 
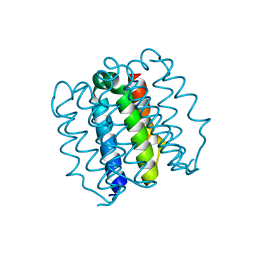 | | Hallucinated C4 protein assembly HALC4_136 | | Descriptor: | HALC4_136 | | Authors: | Ragotte, R.J, Bera, A.K, Wicky, B.I.M, Milles, L.F, Baker, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hallucinating symmetric protein assemblies.
Science, 378, 2022
|
|
8P6H
 
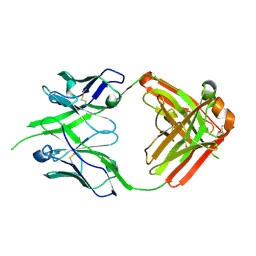 | | Bovine naive ultralong antibody AbBLV5B8* collected at 100K | | Descriptor: | Antibody BLV5B8* heavy chain, Antibody BLV5B8* light chain | | Authors: | Clarke, J.D, Douangamath, A, Mikolajek, H, Stuart, D.I, Owens, R.J. | | Deposit date: | 2023-05-26 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The impact of chain-exchange on bovine ultralong immunoglobulins.
Acta Crystallographica Section F, 2023
|
|
8D06
 
 | | Hallucinated C3 protein assembly HALC3_104 | | Descriptor: | HALC3_104 | | Authors: | Ragotte, R.J, Bera, A.K, Wicky, B.I.M, Milles, L.F, Baker, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Hallucinating symmetric protein assemblies.
Science, 378, 2022
|
|
8P2T
 
 | | Bovine naive ultralong antibody AbD08* collected at 100K | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Antibody D08* heavy chain, Antibody D08* light chain, ... | | Authors: | Clarke, J.D, Douangamath, A, Mikolajek, H, Stuart, D.I, Owens, R.J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The impact of chain-exchange on bovine ultralong immunoglobulins.
Acta Crystallographica Section F, 2024
|
|
1ANB
 
 | |
1AND
 
 | |
8CTB
 
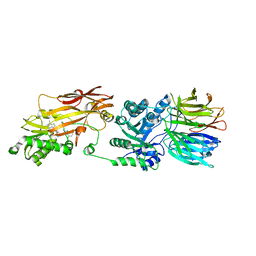 | | Human PRMT5:MEP50 structure with Fragment 3 and MTA Bound | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, 7-chloro-1-methyl-1H-benzimidazol-2-amine, Methylosome protein 50, ... | | Authors: | Gunn, R.J, Lawson, J.D, Smith, C.R. | | Deposit date: | 2022-05-13 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
8OS2
 
 | | Structure of the human LYVE-1 (lymphatic vessel endothelial receptor-1) hyaluronan binding domain in an unliganded state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Lymphatic vessel endothelial hyaluronic acid receptor 1 | | Authors: | Ni, T, Bannerji, S, Jackson, D.J, Gilbert, R.J.C. | | Deposit date: | 2023-04-17 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A novel sliding interaction with hyaluronan underlies the mode of action of LYVE-1, the receptor for leucocyte entry and trafficking in the lymphatics
To be published
|
|
