4Y76
 
 | | Factor Xa complex with GTC000401 | | Descriptor: | CALCIUM ION, Coagulation factor X, N~2~-[(6-chloronaphthalen-2-yl)sulfonyl]-N~2~-{(3S)-1-[(2S)-1-(4-methyl-1,4-diazepan-1-yl)-1-oxopropan-2-yl]-2-oxopyrrolidin-3-yl}glycinamide | | Authors: | Convery, M.A, Young, R.J, Senger, S, Hamblin, J.N, Chan, C, Toomey, J.R, Watson, N.S. | | Deposit date: | 2015-02-13 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Factor Xa inhibitors: S1 binding interactions of a series of N-{(3S)-1-[(1S)-1-methyl-2-morpholin-4-yl-2-oxoethyl]-2-oxopyrrolidin-3-yl}sulfonamides.
J. Med. Chem., 50, 2007
|
|
4Y79
 
 | | Factor Xa complex with GTC000406 | | Descriptor: | (E)-2-(4-chlorophenyl)-N-{(3S)-1-[(2S)-1-(morpholin-4-yl)-1-oxopropan-2-yl]-2-oxopyrrolidin-3-yl}ethenesulfonamide, CALCIUM ION, Coagulation factor X, ... | | Authors: | Convery, M.A, Young, R.J, Senger, S, Hamblin, J.N, Chan, C, Toomey, J.R, Watson, N.S. | | Deposit date: | 2015-02-13 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Factor Xa inhibitors: S1 binding interactions of a series of N-{(3S)-1-[(1S)-1-methyl-2-morpholin-4-yl-2-oxoethyl]-2-oxopyrrolidin-3-yl}sulfonamides.
J. Med. Chem., 50, 2007
|
|
3JR8
 
 | | Crystal Structure of BthTX-II (Asp49-PLA2 from Bothrops jararacussu snake venom) with calcium ions | | Descriptor: | CALCIUM ION, Phospholipase A2 bothropstoxin-2 | | Authors: | Borges, R.J, dos Santos, J.I, Fontes, M.R.M. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural, functional, and bioinformatics studies reveal a new snake venom homologue phospholipase A2 class.
Proteins, 79, 2011
|
|
3J9C
 
 | | CryoEM single particle reconstruction of anthrax toxin protective antigen pore at 2.9 Angstrom resolution | | Descriptor: | CALCIUM ION, Protective antigen PA-63 | | Authors: | Jiang, J, Pentelute, B.L, Collier, R.J, Zhou, Z.H. | | Deposit date: | 2014-12-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Atomic structure of anthrax protective antigen pore elucidates toxin translocation.
Nature, 521, 2015
|
|
4XVU
 
 | | Structure of Get3 bound to the transmembrane domain of Nyv1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, Antibody heavy chain, ... | | Authors: | Mateja, A, Paduch, M, Chang, H.-Y, Szydlowska, A, Kossiakoff, A.A, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2015-01-28 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo.
Science, 347, 2015
|
|
4XTR
 
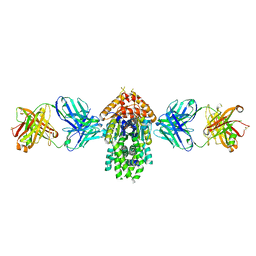 | | Structure of Get3 bound to the transmembrane domain of Pep12 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, ... | | Authors: | Mateja, A, Paduch, M, Chang, H.-Y, Szydlowska, A, Kossiakoff, A.A, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2015-01-23 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo.
Science, 347, 2015
|
|
3K6P
 
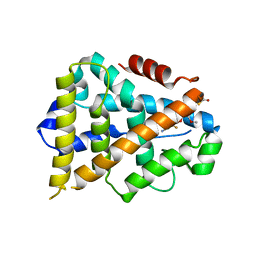 | | Estrogen Related Receptor alpha in Complex with an Ether Based Ligand | | Descriptor: | 4-(4-{[(5R)-2,4-dioxo-1,3-thiazolidin-5-yl]methyl}-2-methoxyphenoxy)-3-(trifluoromethyl)benzonitrile, Steroid hormone receptor ERR1 | | Authors: | Abad, M.C, Patch, R.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Development of Diaryl Ether based ligands for Estrogen Related Receptor alpha as Potential Anti-Diabetic Agents.
To be Published
|
|
6IBK
 
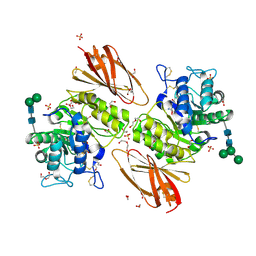 | | Crystal structure of human alpha-galactosidase A in complex with alpha-galactose configured cyclosulfamidate ME763 | | Descriptor: | (3~{a}~{R},4~{S},5~{S},6~{S},7~{R},7~{a}~{S})-7-(hydroxymethyl)-2,2-bis(oxidanylidene)-3~{a},4,5,6,7,7~{a}-hexahydro-3~{H}-benzo[d][1,2,3]oxathiazole-4,5,6-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Wu, L, Davies, G.J. | | Deposit date: | 2018-11-30 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Alpha-d-Gal-cyclophellitol cyclosulfamidate is a Michaelis complex analog that stabilizes therapeutic lysosomal alpha-galactosidase A in Fabry disease
Chem Sci, 2019
|
|
5FK0
 
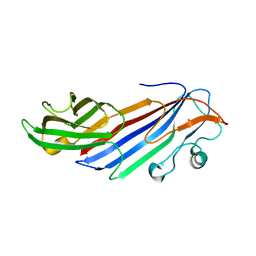 | |
3J35
 
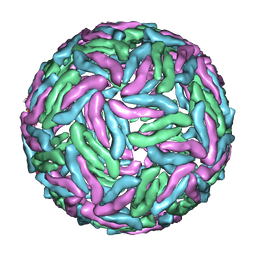 | | Cryo-EM reconstruction of Dengue virus at 37 C | | Descriptor: | envelope protein | | Authors: | Zhang, X.Z, Sheng, J, Plevka, P, Kuhn, R.J, Diamond, M.S, Rossmann, M.G. | | Deposit date: | 2013-02-24 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | Dengue structure differs at the temperatures of its human and mosquito hosts.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1VZS
 
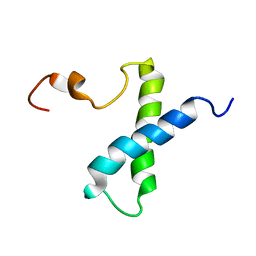 | | Solution structure of subunit F6 from the peripheral stalk region of ATP synthase from bovine heart mitochondria | | Descriptor: | ATP SYNTHASE COUPLING FACTOR 6, MITOCHONDRIAL PRECURSOR | | Authors: | Carbajo, R.J, Silvester, J.A, Runswick, M.J, Walker, J.E, Neuhaus, D. | | Deposit date: | 2004-05-25 | | Release date: | 2004-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Subunit F(6) from the Peripheral Stalk Region of ATP Synthase from Bovine Heart Mitochondria
J.Mol.Biol., 342, 2004
|
|
6I7L
 
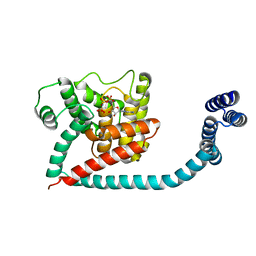 | | Crystal structure of monomeric FICD mutant L258D complexed with MgAMP-PNP | | Descriptor: | Adenosine monophosphate-protein transferase FICD, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
5HK5
 
 | |
1UWA
 
 | | L290F mutant rubisco from chlamydomonas | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Karkehabadi, S, Taylor, T.C, Spreitzer, R.J, Andersson, I. | | Deposit date: | 2004-02-03 | | Release date: | 2005-01-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Altered Intersubunit Interactions in Crystal Structures of Catalytically Compromised Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase
Biochemistry, 44, 2005
|
|
6I7G
 
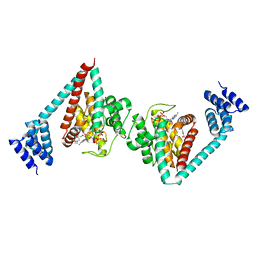 | | Crystal structure of dimeric wild type FICD complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenosine monophosphate-protein transferase FICD, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
5IOM
 
 | | Crystal Structure of Nucleoside Diphosphate Kinase from Schistosoma mansoni is space group P6322 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Torini, J.R.S, Romanello, L, Bird, L.E, Nettleship, J.E, Owens, R.J, Aller, P, DeMarco, R, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2016-03-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of a Schistosoma mansoni NDPK expressed in sexual and digestive organs.
Mol.Biochem.Parasitol., 2019
|
|
1Y8M
 
 | |
1YFI
 
 | | Crystal Structure of restriction endonuclease MspI in complex with its cognate DNA in P212121 space group | | Descriptor: | 5'-D(*CP*CP*CP*CP*CP*GP*GP*GP*GP*G)-3', Type II restriction enzyme MspI | | Authors: | Xu, Q.S, Kucera, R.B, Roberts, R.J, Guo, H.-C. | | Deposit date: | 2005-01-02 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two crystal forms of the restriction enzyme MspI-DNA complex show the same novel structure.
Protein Sci., 14, 2005
|
|
4ZHC
 
 | | Siderocalin-mediated recognition and cellular uptake of actinides | | Descriptor: | ACETATE ION, N-{2-[bis(2-{[(2,3-dihydroxyphenyl)carbonyl]amino}ethyl)amino]ethyl}-1-hydroxy-6-oxo-1,6-dihydropyridine-2-carboxamide, Neutrophil gelatinase-associated lipocalin, ... | | Authors: | Allred, B.E, Rupert, P.B, Gauny, S.S, An, D.D, Ralston, C.Y, Sturzbecher-Hoehne, M, Strong, R.K, Abergel, R.J. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Siderocalin-mediated recognition, sensitization, and cellular uptake of actinides.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZHO
 
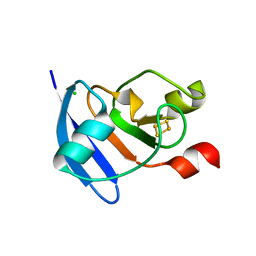 | | The crystal structure of Arabidopsis ferredoxin 2 with 2Fe-2S cluster | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-2, ... | | Authors: | Grinter, R, Josts, I, Roszak, A.W, Cogdell, R.J, Walker, D. | | Deposit date: | 2015-04-26 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure of the bacterial plant-ferredoxin receptor FusA.
Nat Commun, 7, 2016
|
|
4ZHF
 
 | | Siderocalin-mediated recognition and cellular uptake of actinides | | Descriptor: | CURIUM ION, GLYCEROL, N,N'-butane-1,4-diylbis[1-hydroxy-N-(3-{[(1-hydroxy-6-oxo-1,6-dihydropyridin-2-yl)carbonyl]amino}propyl)-6-oxo-1,6-dihydropyridine-2-carboxamide], ... | | Authors: | Allred, B.E, Rupert, P.B, Gauny, S.S, An, D.D, Ralston, C.Y, Sturzbecher-Hoehne, M, Strong, R.K, Abergel, R.J. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Siderocalin-mediated recognition, sensitization, and cellular uptake of actinides.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1YKS
 
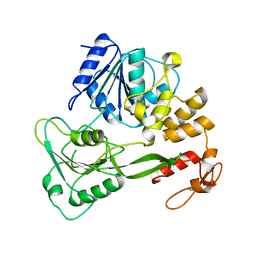 | | Crystal structure of yellow fever virus NS3 helicase | | Descriptor: | Genome polyprotein [contains: Flavivirin protease NS3 catalytic subunit] | | Authors: | Wu, J, Bera, A.K, Kuhn, R.J, Smith, J.L. | | Deposit date: | 2005-01-18 | | Release date: | 2005-08-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the flavivirus helicase: implications for catalytic activity, protein interactions, and proteolytic processing.
J.Virol., 79, 2005
|
|
1VYX
 
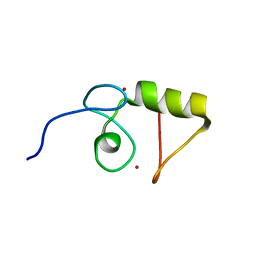 | | Solution structure of the KSHV K3 N-terminal domain | | Descriptor: | ORF K3, ZINC ION | | Authors: | Dodd, R.B, Allen, M.D, Brown, S.E, Sanderson, C.M, Duncan, L.M, lehner, P.J, Bycroft, M, Read, R.J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-10-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Kaposi'S Sarcoma-Associated Herpesvirus K3 N-Terminal Domain Reveals a Novel E2-Binding C4Hc3-Type Ring Domain
J.Biol.Chem., 279, 2004
|
|
3KXA
 
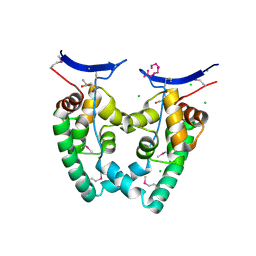 | | Crystal Structure of NGO0477 from Neisseria gonorrhoeae | | Descriptor: | ASPARAGINE, CHLORIDE ION, Putative uncharacterized protein, ... | | Authors: | Ren, J, Sainsbury, S, Nettleship, J.E, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2009-12-02 | | Release date: | 2010-01-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of NGO0477 from Neisseria gonorrhoeae reveals a novel protein fold incorporating a helix-turn-helix motif.
Proteins, 78, 2010
|
|
1VKG
 
 | | Crystal Structure of Human HDAC8 complexed with CRA-19156 | | Descriptor: | 5-(4-METHYL-BENZOYLAMINO)-BIPHENYL-3,4'-DICARBOXYLIC ACID 3-DIMETHYLAMIDE-4'-HYDROXYAMIDE, Histone deacetylase 8, SODIUM ION, ... | | Authors: | Somoza, J.R, Skene, R.J, Katz, B.A, Mol, C, Ho, J.D, Jennings, A.J, Luong, C, Arvai, A, Buggy, J.J, Chi, E, Tang, J, Sang, B.-C, Verner, E, Wynands, R, Leahy, E.M, Dougan, D.R, Snell, G, Navre, M, Knuth, M.W, Swanson, R.V, McRee, D.E, Tari, L.W. | | Deposit date: | 2004-05-13 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Snapshots of Human HDAC8 Provide Insights into the Class I Histone Deacetylases
Structure, 12, 2004
|
|
