5FOG
 
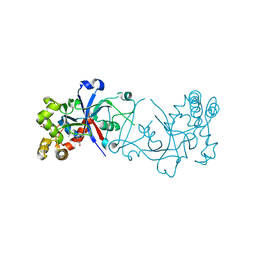 | | Crystal structure of hte Cryptosporidium muris cytosolic leucyl-tRNA synthetase editing domain complex with a post-transfer editing analogue of norvaline (Nv2AA) | | Descriptor: | 1,2-ETHANEDIOL, 2'-(L-NORVALYL)AMINO-2'-DEOXYADENOSINE, LEUCYL-TRNA SYNTHETASE, ... | | Authors: | Palencia, A, Liu, R.J, Lukarska, M, Gut, J, Bougdour, A, Touquet, B, Wang, E.D, Alley, M.R.K, Rosenthal, P.J, Hakimi, M.A, Cusack, S. | | Deposit date: | 2015-11-20 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cryptosporidium and Toxoplasma Parasites are Inhibited by a Benzoxaborole Targeting Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
5FMW
 
 | | The poly-C9 component of the Complement Membrane Attack Complex | | Descriptor: | POLYC9 | | Authors: | Dudkina, N.V, Spicer, B.A, Reboul, C.F, Conroy, P.J, Lukoyanova, N, Elmlund, H, Law, R.H.P, Ekkel, S.M, Kondos, S.C, Goode, R.J.A, Ramm, G, Whisstock, J.C, Saibil, H.R, Dunstone, M.A. | | Deposit date: | 2015-11-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of the Poly-C9 Component of the Complement Membrane Attack Complex
Nat.Commun., 7, 2016
|
|
5G21
 
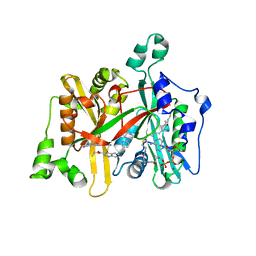 | | Leishmania major N-myristoyltransferase in complex with a quinoline inhibitor (compound 26). | | Descriptor: | ETHYL 4-[(2-CYANOETHYL)SULFANYL]-6-{[6-(PIPERAZIN-1-YL), GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Goncalves, V, Brannigan, J.A, Laporte, A, Bell, A.S, Roberts, S.M, Wilkinson, A.J, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2016-04-06 | | Release date: | 2017-02-15 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided optimization of quinoline inhibitors of Plasmodium N-myristoyltransferase.
Medchemcomm, 8, 2017
|
|
5G1Z
 
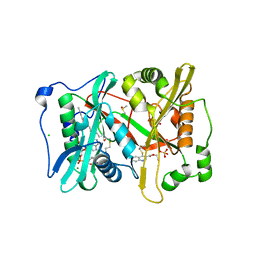 | | Plasmodium vivax N-myristoyltransferase in complex with a quinoline inhibitor (compound 1) | | Descriptor: | 2-oxopentadecyl-CoA, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Goncalves, V, Brannigan, J.A, Laporte, A, Bell, A.S, Roberts, S.M, Wilkinson, A.J, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2016-04-06 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided optimization of quinoline inhibitors of Plasmodium N-myristoyltransferase.
Medchemcomm, 8, 2017
|
|
3EI0
 
 | |
6U01
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, N84D mutant with pyruvate bound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Lehnert, L, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2019-08-13 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J.Struct.Biol., 209, 2020
|
|
6TZU
 
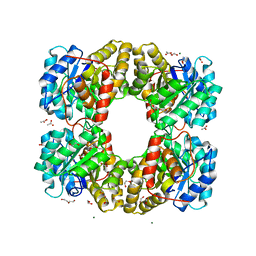 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, N84A mutant with pyruvate bound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Lehnert, C, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2019-08-13 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J.Struct.Biol., 209, 2020
|
|
1PGV
 
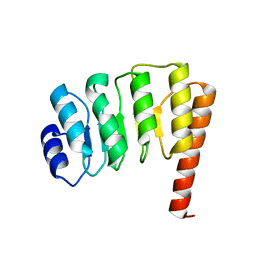 | | Structural Genomics of Caenorhabditis elegans: tropomodulin C-terminal domain | | Descriptor: | tropomodulin TMD-1 | | Authors: | Symersky, J, Lu, S, Li, S, Chen, L, Meehan, E, Luo, M, Qiu, S, Bunzel, R.J, Luo, D, Arabashi, A, Nagy, L.A, Lin, G, Luan, W.C.-H, Carson, M, Gray, R, Huang, W, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-05-28 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: crystal structure of the tropomodulin C-terminal domain
Proteins, 56, 2004
|
|
1YJD
 
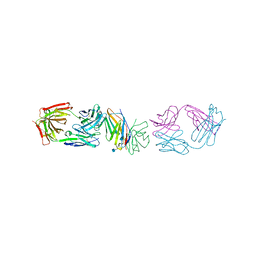 | | Crystal structure of human CD28 in complex with the Fab fragment of a mitogenic antibody (5.11A1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab fragment of 5.11A1 antibody heavy chain, Fab fragment of 5.11A1 antibody light chain, ... | | Authors: | Evans, E.J, Esnouf, R.M, Manso-Sancho, R, Gilbert, R.J.C, James, J.R, Sorensen, P, Stuart, D.I, Davis, S.J. | | Deposit date: | 2005-01-14 | | Release date: | 2005-02-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a soluble CD28-Fab complex
Nat.Immunol., 6, 2005
|
|
1SRK
 
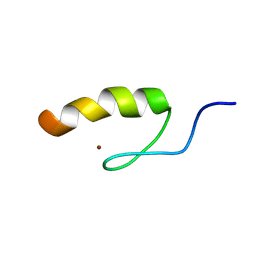 | | Solution structure of the third zinc finger domain of FOG-1 | | Descriptor: | ZINC ION, Zinc finger protein ZFPM1 | | Authors: | Simpson, R.J.Y, Lee, S.H.Y, Bartle, N, Matthews, J.M, Mackay, J.P, Crossley, M. | | Deposit date: | 2004-03-22 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A Classic Zinc Finger from Friend of GATA Mediates an Interaction with the Coiled-coil of Transforming Acidic Coiled-coil 3.
J.Biol.Chem., 279, 2004
|
|
3EHZ
 
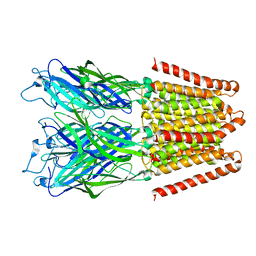 | |
8X7T
 
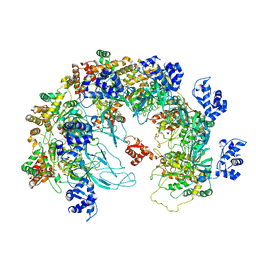 | | MCM in the Apo state. | | Descriptor: | mini-chromosome maintenance complex 3 | | Authors: | Ma, J, Yi, G, Ye, M, MacGregor-Chatwin, C, Sheng, Y, Lu, Y, Li, M, Gilbert, R.J.C, Zhang, P. | | Deposit date: | 2023-11-25 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | MCM in the Apo state
To Be Published
|
|
8X7U
 
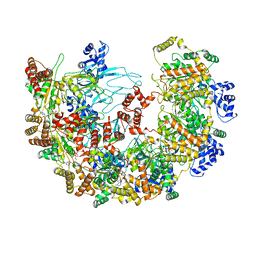 | | MCM in complex with dsDNA in presence of ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, mini-chromosome maintenance complex 3 | | Authors: | Ma, J, Yi, G, Ye, M, MacGregor-Chatwin, C, Sheng, Y, Lu, Y, Li, M, Gilbert, R.J.C, Zhang, P. | | Deposit date: | 2023-11-25 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | MCM in complex with dsDNA in presence of ATP
To Be Published
|
|
1Y0J
 
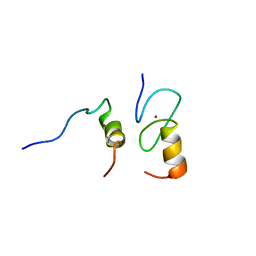 | | Zinc fingers as protein recognition motifs: structural basis for the GATA-1/Friend of GATA interaction | | Descriptor: | Erythroid transcription factor, ZINC ION, Zinc-finger protein ush | | Authors: | Liew, C.K, Simpson, R.J.Y, Kwan, A.H.Y, Crofts, L.A, Loughlin, F.E, Matthews, J.M, Crossley, M, Mackay, J.P. | | Deposit date: | 2004-11-15 | | Release date: | 2005-01-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Zinc fingers as protein recognition motifs: Structural basis for the GATA-1/Friend of GATA interaction
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1SQN
 
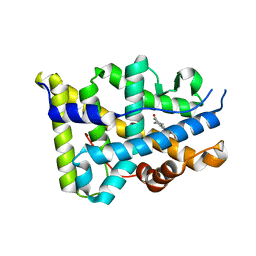 | | Progesterone Receptor Ligand Binding Domain with bound Norethindrone | | Descriptor: | (14beta,17alpha)-17-ethynyl-17-hydroxyestr-4-en-3-one, progesterone receptor | | Authors: | Williams, S.P, Madauss, K.P, Deng, J.-S, Austin, R.J.H, Lambert, M.H, McLay, I, Pritchard, J, Short, S.A, Stewart, E.L, Uings, I.J. | | Deposit date: | 2004-03-19 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Progesterone receptor ligand binding pocket flexibility: crystal structures of the norethindrone and mometasone furoate complexes
J.Med.Chem., 47, 2004
|
|
2L4K
 
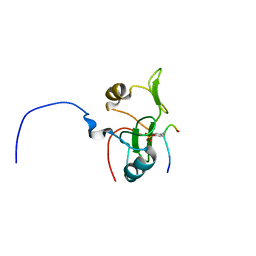 | | Water refined solution structure of the human Grb7-SH2 domain in complex with the 10 amino acid peptide pY1139 | | Descriptor: | Growth factor receptor-bound protein 7, Receptor tyrosine-protein kinase erbB-2 | | Authors: | Pias, S.C, Ivancic, M, Brescia, P.J, Johnson, D.L, Smith, D.E, Daly, R.J, Lyons, B.A. | | Deposit date: | 2010-10-07 | | Release date: | 2010-11-17 | | Last modified: | 2012-10-03 | | Method: | SOLUTION NMR | | Cite: | Water-Refined Solution Structure of the Human Grb7-SH2 Domain in Complex with the erbB2 Receptor Peptide pY1139.
Protein Pept.Lett., 19, 2012
|
|
5UF4
 
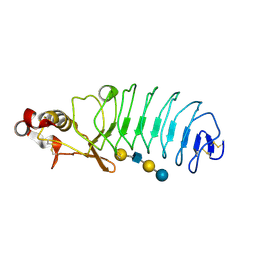 | | Crystal Structure of Variable Lymphocyte Receptor (VLR) O13 with LNnT bound | | Descriptor: | O13, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Gunn, R.J, Collins, B.C, McKitrick, T.R, Cummings, R.D, Cooper, M.D, Herrin, B.R, Wilson, I.A. | | Deposit date: | 2017-01-03 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Insights into VLR Fine Specificity for Blood Group Carbohydrates.
Structure, 25, 2017
|
|
5UFB
 
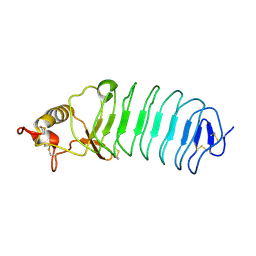 | | Crystal Structure of Variable Lymphocyte Receptor (VLR) Tn4-22 (Apo) | | Descriptor: | Tn4-22 | | Authors: | Collins, B.C, Gunn, R.J, McKitrick, T.R, Cummings, R.D, Cooper, M.D, Herrin, B.R, Wilson, I.A. | | Deposit date: | 2017-01-04 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Structural Insights into VLR Fine Specificity for Blood Group Carbohydrates.
Structure, 25, 2017
|
|
5UFF
 
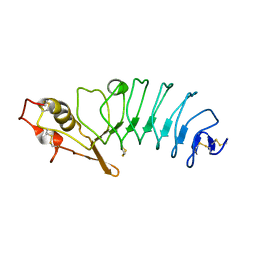 | | Crystal Structure of Variable Lymphocyte Receptor (VLR) RBC36 with Fucose(alpha-1-2)Lactose bound | | Descriptor: | RBC36, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Collins, B.C, Gunn, R.J, McKitrick, T.R, Herrin, B.R, Cummings, R.D, Cooper, M.D, Wilson, I.A. | | Deposit date: | 2017-01-04 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | Structural Insights into VLR Fine Specificity for Blood Group Carbohydrates.
Structure, 25, 2017
|
|
5U75
 
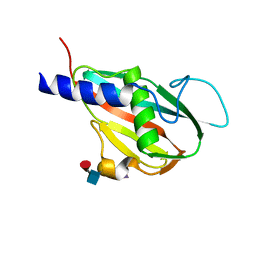 | | The structure of Staphylococcal Enterotoxin-like X (SElX), a Unique Superantigen | | Descriptor: | Enterotoxin-like toxin X, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Ting, Y.T, Young, P.G, Langley, R.J, Baker, H, Fraser, J.D. | | Deposit date: | 2016-12-11 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Staphylococcal enterotoxin-like X (SElX) is a unique superantigen with functional features of two major families of staphylococcal virulence factors.
PLoS Pathog., 13, 2017
|
|
5T1A
 
 | | Structure of CC Chemokine Receptor 2 with Orthosteric and Allosteric Antagonists | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{R})-1-(4-chloranyl-2-fluoranyl-phenyl)-2-cyclohexyl-3-ethanoyl-4-oxidanyl-2~{H}-pyrrol-5-one, (3S)-1-{(1S,2R,4R)-4-[methyl(propan-2-yl)amino]-2-propylcyclohexyl}-3-{[6-(trifluoromethyl)quinazolin-4-yl]amino}pyrrolidin-2-one, ... | | Authors: | Zheng, Y, Qin, L, Ortiz Zacarias, N.V, de Vries, H, Han, G.W, Gustavsson, M, Dabros, M, Zhao, C, Cherney, R.J, Carter, P, Stamos, D, Abagyan, R, Cherezov, V, Stevens, R.C, IJzerman, A.P, Heitman, L.H, Tebben, A, Kufareva, I, Handel, T.M. | | Deposit date: | 2016-08-18 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists.
Nature, 540, 2016
|
|
5S7F
 
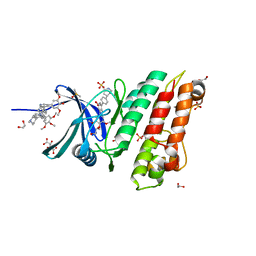 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM010935a | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, Activin receptor type-1, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S7O
 
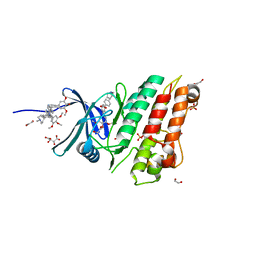 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM007391c | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, Activin receptor type-1, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S7R
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM010918a | | Descriptor: | 1,2-ETHANEDIOL, 1lambda~6~,2-thiazetidine-1,1-dione, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S80
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM010946a | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, Activin receptor type-1, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | XChem group deposition
To Be Published
|
|
