6DO1
 
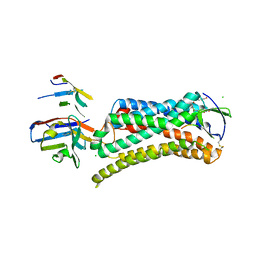 | | Structure of nanobody-stabilized angiotensin II type 1 receptor bound to S1I8 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-like peptide S1I8, ... | | Authors: | Wingler, L.M, McMahon, C, Staus, D.P, Lefkowitz, R.J, Kruse, A.C. | | Deposit date: | 2018-06-08 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Distinctive Activation Mechanism for Angiotensin Receptor Revealed by a Synthetic Nanobody.
Cell, 176, 2019
|
|
6DQL
 
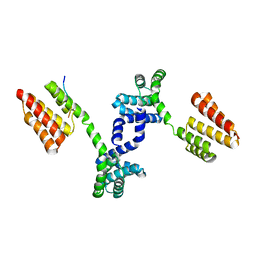 | | Crystal structure of Regulator of Proteinase B RopB complexed with SIP | | Descriptor: | Regulator of Proteinase B RopB, SpeB-inducing peptide (SIP) | | Authors: | Do, H, Makthal, N, VanderWal, A.R, Olsen, R.J, Musser, J.M, Kumaraswami, M. | | Deposit date: | 2018-06-11 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Environmental pH and peptide signaling control virulence of Streptococcus pyogenes via a quorum-sensing pathway.
Nat Commun, 10, 2019
|
|
4PPK
 
 | | Crystal structure of eCGP123 T69V variant at pH 7.5 | | Descriptor: | Monomeric Azami Green | | Authors: | Don Paul, C, Traore, D.A.K, Devenish, R.J, Close, D, Bell, T, Bradbury, A, Wilce, M.C.J, Prescott, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Crystal Structure and Properties of Phanta, a Weakly Fluorescent Photochromic GFP-Like Protein.
Plos One, 10, 2015
|
|
6DDA
 
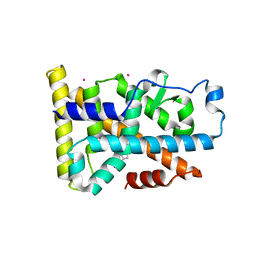 | | Nurr1 Covalently Modified by a Dopamine Metabolite | | Descriptor: | 5-hydroxy-1,2-dihydro-6H-indol-6-one, BROMIDE ION, Nuclear receptor subfamily 4 group A member 2, ... | | Authors: | Bruning, J.M, Wang, Y, Otrabella, F, Boxue, T, Liu, H, Bhattacharya, P, Guo, S, Holton, J.M, Fletterick, R.J, Jacobson, M.P, England, P.M. | | Deposit date: | 2018-05-09 | | Release date: | 2019-03-20 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Covalent Modification and Regulation of the Nuclear Receptor Nurr1 by a Dopamine Metabolite.
Cell Chem Biol, 26, 2019
|
|
4P8S
 
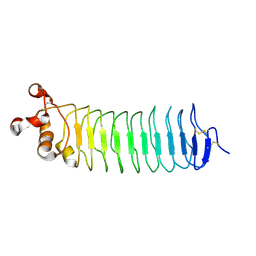 | | Crystal structure of Nogo-receptor-2 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Semavina, M, Saha, N, Kolev, M.V, Giger, R.J, Himanen, J.P, Nikolov, D.B. | | Deposit date: | 2014-04-01 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Nogo-receptor-2.
Protein Sci., 2011
|
|
3CL5
 
 | | Structure of coronavirus hemagglutinin-esterase in complex with 4,9-O-diacetyl sialic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Zeng, Q.H, Langereis, M.A, van Vliet, A.L.W, Huizinga, E.G, de Groot, R.J. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
8RVO
 
 | | Proteasomal late precursor complex from pre1-1, state 1 | | Descriptor: | Probable proteasome subunit alpha type-7, Proteasome assembly chaperone 2, Proteasome chaperone 1, ... | | Authors: | Mark, E, Ramos, P.C, Kayser, F, Hoeckendorff, J, Dohmen, R.J, Wendler, P. | | Deposit date: | 2024-02-01 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural roles of Ump1 and beta-subunit propeptides in proteasome biogenesis.
Life Sci Alliance, 7, 2024
|
|
4PPL
 
 | | Crystal structure of eCGP123 H193Q variant at pH 7.5 | | Descriptor: | Monomeric Azami Green | | Authors: | Don Paul, C, Traore, D.A.K, Devenish, R.J, Close, D, Bell, T, Bradbury, A, Wilce, M.C.J, Prescott, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray Crystal Structure and Properties of Phanta, a Weakly Fluorescent Photochromic GFP-Like Protein.
Plos One, 10, 2015
|
|
6DGM
 
 | | Streptococcus pyogenes phosphoglycerol transferase GacH in complex with sn-glycerol-1-phosphate | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, Phosphoglycerol transferase GacH, ... | | Authors: | Edgar, R.J, Korotkova, N, Korotkov, K.V. | | Deposit date: | 2018-05-17 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of glycerol phosphate modification on streptococcal rhamnose polysaccharides.
Nat.Chem.Biol., 15, 2019
|
|
8RVP
 
 | | Proteasomal late precursor complex from pre1-1, state 2 | | Descriptor: | Probable proteasome subunit alpha type-7, Proteasome assembly chaperone 2, Proteasome chaperone 1, ... | | Authors: | Mark, E, Ramos, P.C, Kayser, F, Hoeckendorff, J, Dohmen, R.J, Wendler, P. | | Deposit date: | 2024-02-01 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.28 Å) | | Cite: | Structural roles of Ump1 and beta-subunit propeptides in proteasome biogenesis.
Life Sci Alliance, 7, 2024
|
|
8RVQ
 
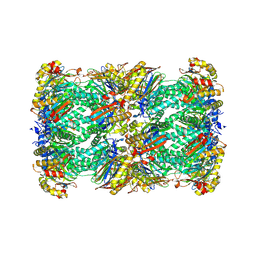 | | 20S proteasome from pre1-1 | | Descriptor: | Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Mark, E, Ramos, P.C, Kayser, F, Hoeckendorff, J, Dohmen, R.J, Wendler, P. | | Deposit date: | 2024-02-01 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.02 Å) | | Cite: | Structural roles of Ump1 and beta-subunit propeptides in proteasome biogenesis.
Life Sci Alliance, 7, 2024
|
|
4PK9
 
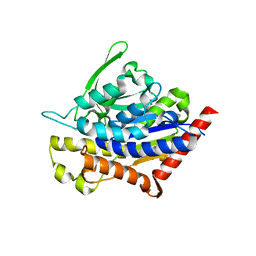 | |
8RVL
 
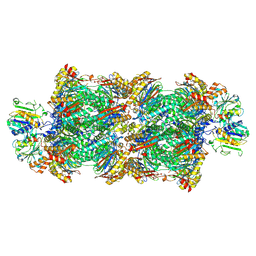 | | Proteasomal late precursor complex from pre1-1 | | Descriptor: | Probable proteasome subunit alpha type-7, Proteasome assembly chaperone 2, Proteasome chaperone 1, ... | | Authors: | Mark, E, Ramos, P.C, Kayser, F, Hoeckendorff, J, Dohmen, R.J, Wendler, P. | | Deposit date: | 2024-02-01 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Structural roles of Ump1 and beta-subunit propeptides in proteasome biogenesis.
Life Sci Alliance, 7, 2024
|
|
5EG2
 
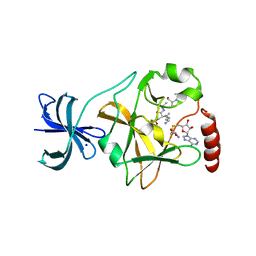 | | SET7/9 N265A in complex with AdoHcy and TAF10 peptide | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, ... | | Authors: | Kroner, G.M, Fick, R.J, Trievel, R.C. | | Deposit date: | 2015-10-26 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Sulfur-Oxygen Chalcogen Bonding Mediates AdoMet Recognition in the Lysine Methyltransferase SET7/9.
Acs Chem.Biol., 11, 2016
|
|
3CL2
 
 | | N1 Neuraminidase N294S + Oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, Neuraminidase | | Authors: | Collins, P, Haire, L.F, Lin, Y.P, Liu, J, Russell, R.J, Walker, P.A, Skehel, J.J, Martin, S.R, Hay, A.J, Gamblin, S.J. | | Deposit date: | 2008-03-18 | | Release date: | 2008-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.538 Å) | | Cite: | Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants.
Nature, 453, 2008
|
|
3COB
 
 | | Structural Dynamics of the Microtubule binding and regulatory elements in the Kinesin-like Calmodulin binding protein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin heavy chain-like protein, MAGNESIUM ION | | Authors: | Vinogradova, M.V, Malanina, G.G, Reddy, V, Reddy, A.S.N, Fletterick, R.J. | | Deposit date: | 2008-03-27 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural dynamics of the microtubule binding and regulatory elements in the kinesin-like calmodulin binding protein.
J.Struct.Biol., 163, 2008
|
|
3IJY
 
 | | Structure of S67-27 in Complex with Kdo(2.8)Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, Immunoglobulin heavy chain (IGG3), Immunoglobulin light chain (IGG3), ... | | Authors: | Brooks, C.L, Blackler, R.J, Evans, S.V. | | Deposit date: | 2009-08-05 | | Release date: | 2009-10-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The role of CDR H3 in antibody recognition of a synthetic analog of a lipopolysaccharide antigen.
Glycobiology, 20, 2010
|
|
8RLT
 
 | |
8RLU
 
 | |
8RLV
 
 | |
4Q4E
 
 | | Crystal structure of E.coli aminopeptidase N in complex with actinonin | | Descriptor: | ACTINONIN, Aminopeptidase N, GLYCEROL, ... | | Authors: | Reddi, R, Ganji, R.J, Addlagatta, A. | | Deposit date: | 2014-04-14 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the inhibition of M1 family aminopeptidases by the natural product actinonin: Crystal structure in complex with E. coli aminopeptidase N.
Protein Sci., 24, 2015
|
|
6E0I
 
 | | Crystal structure of Glucokinase in complex with compound 72 | | Descriptor: | 1-{4-[5-({3-[(2-methylpyridin-3-yl)oxy]-5-[(pyridin-2-yl)sulfanyl]pyridin-2-yl}amino)-1,2,4-thiadiazol-3-yl]piperidin-1 -yl}ethan-1-one, DIMETHYL SULFOXIDE, Glucokinase, ... | | Authors: | Hinklin, R.J, Baer, B.R, Boyd, S.A, Chicarelli, M.D, Condroski, K.R, DeWolf, W.E, Fischer, J, Frank, M, Hingorani, G.P, Lee, P.A, Neitzel, N.A, Pratt, S.A, Singh, A, Sullivan, F.X, Turner, T, Voegtli, W.C, Wallace, E.M, Williams, L, Aicher, T.D. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and preclinical development of AR453588 as an anti-diabetic glucokinase activator.
Bioorg.Med.Chem., 28, 2020
|
|
9FTI
 
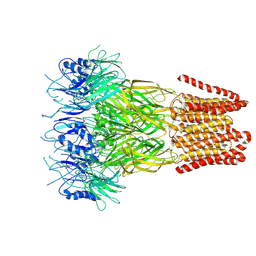 | | Open conformation of the pentameric ligand-gated ion channel, DeCLIC at pH 5 with 10mM EDTA | | Descriptor: | Neurotransmitter-gated ion-channel ligand-binding domain-containing protein | | Authors: | Rovsnik, U, Anden, O, Lycksell, M, Delarue, M, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-06-24 | | Release date: | 2024-09-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural characterization of pH-modulated closed and open states
in a pentameric ligand-gated ion channel
To Be Published
|
|
9GBK
 
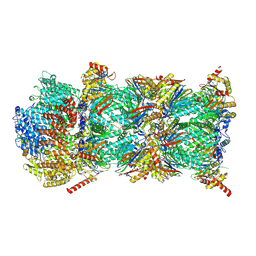 | | Blm10-20S proteasome complex from pre1-1 | | Descriptor: | Probable proteasome subunit alpha type-7, Proteasome activator BLM10, Proteasome subunit alpha type-1, ... | | Authors: | Mark, E, Ramos, P.C, Kayser, F, Hoeckendorff, J, Dohmen, R.J, Wendler, P. | | Deposit date: | 2024-07-31 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Structural roles of Ump1 and beta-subunit propeptides in proteasome biogenesis.
Life Sci Alliance, 7, 2024
|
|
9FTG
 
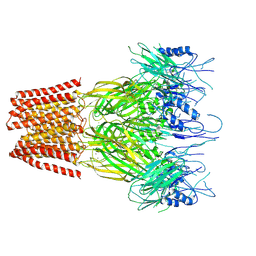 | | Closed conformation of the pentameric ligand-gated ion channel, DeCLIC at pH 5 with 10 mM Ca2+ | | Descriptor: | CALCIUM ION, Neurotransmitter-gated ion-channel ligand-binding domain-containing protein | | Authors: | Rovsnik, U, Anden, O, Lycksell, M, Delarue, M, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-06-24 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural characterization of pH-modulated closed and open states
in a pentameric ligand-gated ion channel
To Be Published
|
|
