8G4L
 
 | | Cryo-EM structure of the human cardiac myosin filament | | Descriptor: | Myosin light chain 3, Myosin regulatory light chain 2, ventricular/cardiac muscle isoform, ... | | Authors: | Dutta, D, Nguyen, V, Padron, R, Craig, R. | | Deposit date: | 2023-02-10 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Cryo-EM structure of the human cardiac myosin filament.
Nature, 623, 2023
|
|
6OAU
 
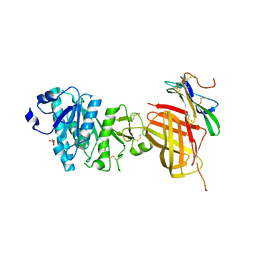 | | Apo Structure of WT Lipoprotein Lipase in Complex with GPIHBP1 Mutant N78D N82D produced in GnTI-deficient HEK293-F cells | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1, ... | | Authors: | Arora, R, Horton, P.A, Benson, T.E, Romanowski, M.J. | | Deposit date: | 2019-03-18 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure of lipoprotein lipase in complex with GPIHBP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OB0
 
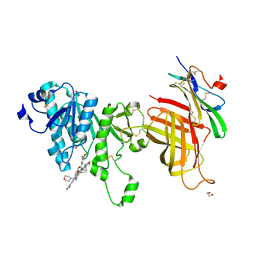 | | Compound 2 bound structure of WT Lipoprotein Lipase in Complex with GPIHBP1 Mutant N78D N82D produced in HEK293-F cells | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Arora, R, Horton, P.A, Benson, T.E, Romanowski, M.J. | | Deposit date: | 2019-03-19 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure of lipoprotein lipase in complex with GPIHBP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7P1K
 
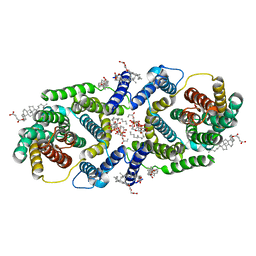 | | Cryo EM structure of bison NHA2 in nano disc structure | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Phosphatidylinositol, mitochondrial sodium/hydrogen exchanger 9B2 | | Authors: | Matsuoka, R, Fudim, R, Jung, S, Drew, D. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-26 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structure, mechanism and lipid-mediated remodeling of the mammalian Na + /H + exchanger NHA2.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P1J
 
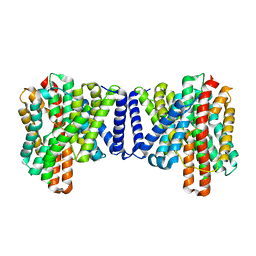 | | Cryo EM structure of bison NHA2 in detergent structure | | Descriptor: | mitochondrial sodium/hydrogen exchanger 9B2 | | Authors: | Matsuoka, R, Fudim, R, Jung, S, Drew, D. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-26 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structure, mechanism and lipid-mediated remodeling of the mammalian Na + /H + exchanger NHA2.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P1I
 
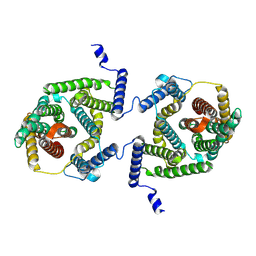 | | Cryo EM structure of bison NHA2 in detergent and N-terminal extension helix | | Descriptor: | mitochondrial sodium/hydrogen exchanger 9B2 | | Authors: | Matsuoka, R, Fudim, R, Jung, S, Drew, D. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-26 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structure, mechanism and lipid-mediated remodeling of the mammalian Na + /H + exchanger NHA2.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7A3F
 
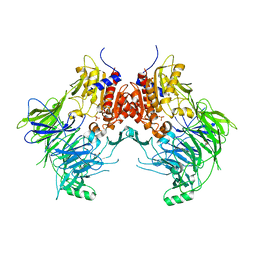 | | Crystal structure of apo DPP9 | | Descriptor: | Dipeptidyl peptidase 9, GLYCEROL, PHOSPHATE ION | | Authors: | Ross, B.H, Huber, R. | | Deposit date: | 2020-08-18 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery and Development of 4-Oxo-beta-Lactams as Novel Inhibitors of Dipeptidyl Peptidases 8 and 9
To Be Published
|
|
7A3I
 
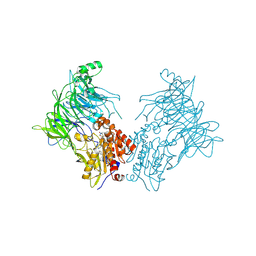 | |
7A3K
 
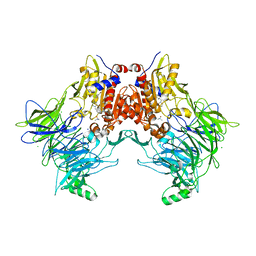 | |
6OAZ
 
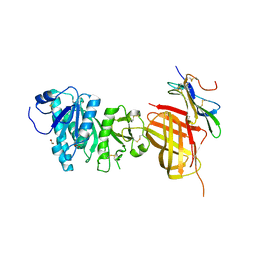 | | Apo Structure of WT Lipoprotein Lipase in Complex with GPIHBP1 Mutant N78D N82D produced in HEK293-F cells | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Arora, R, Horton, P.A, Benson, T.E, Romanowski, M.J. | | Deposit date: | 2019-03-19 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure of lipoprotein lipase in complex with GPIHBP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7B83
 
 | | Structure of SARS-CoV-2 Main Protease bound to pyrithione zinc | | Descriptor: | 3C-like proteinase, 9-oxa-7-thia-1-azonia-8$l^{2}-zincabicyclo[4.3.0]nona-1,3,5-triene, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6ZP7
 
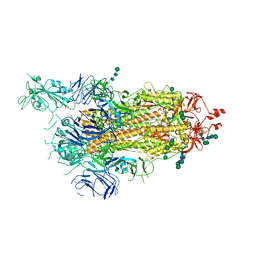 | | SARS-CoV-2 spike in prefusion state (flexibility analysis, 1-up open conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Martinez, M, Marabini, R, Carazo, J.M. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-29 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Continuous flexibility analysis of SARS-CoV-2 spike prefusion structures.
Iucrj, 7, 2020
|
|
6ZOW
 
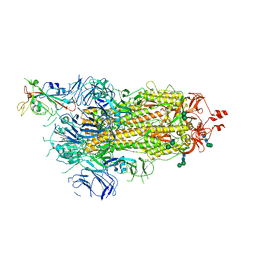 | | SARS-CoV-2 spike in prefusion state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Martinez, M, Marabini, R, Carazo, J.M. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-29 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Continuous flexibility analysis of SARS-CoV-2 spike prefusion structures.
Iucrj, 7, 2020
|
|
6ZP5
 
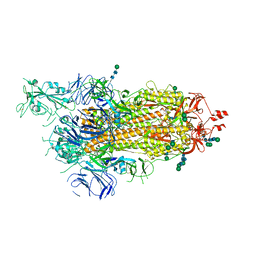 | | SARS-CoV-2 spike in prefusion state (flexibility analysis, 1-up closed conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Martinez, M, Marabini, R, Carazo, J.M. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-29 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Continuous flexibility analysis of SARS-CoV-2 spike prefusion structures.
Iucrj, 7, 2020
|
|
8I7E
 
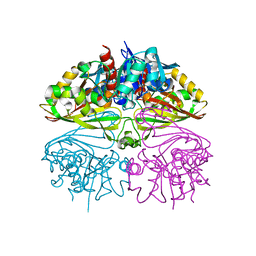 | | Crystal structure of Glyceraldehyde 3-phosphate dehydrogenase from Salmonella typhi at 2.05A | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Kumar, N, Dilawari, R, Chaubey, G.K, Modanwal, R, Talukdar, S, Dhiman, A, Chaudhary, S, Patidar, A, Kumar, A, Raje, C.I, Raje, M, Kumaran, S. | | Deposit date: | 2023-01-31 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Glyceraldehyde 3-phosphate dehydrogenase from Salmonella typhi at 2.05A
To Be Published
|
|
8I7P
 
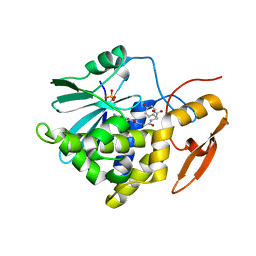 | | Crystal structure of Ricin A chain bound with N2-(2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)glycyl-L-tyrosine | | Descriptor: | 6-(2-ethyl-4-hydroxyphenyl)-1H-indazole-3-carboxamide, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Sakamoto, N, Higashi, S, Kawata, R, Nagatsu, K, Saito, R. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of ricin toxin A chain complexed with a highly potent pterin-based small-molecular inhibitor.
J Enzyme Inhib Med Chem, 38, 2023
|
|
8I9L
 
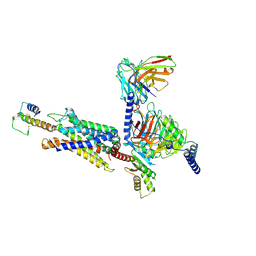 | | Structure of C3a-C3aR-Go complex (Composite map) | | Descriptor: | Antibody fragment - ScFv16, C3a anaphylatoxin, C3a anaphylatoxin chemotactic receptor, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Sarma, P, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-02-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
8I95
 
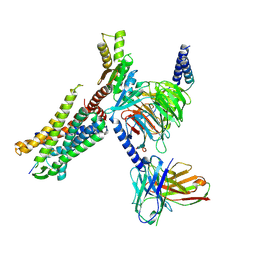 | | Structure of EP54-C3aR-Go complex | | Descriptor: | Antibody fragment - ScFv16, C3a anaphylatoxin chemotactic receptor, EP54 ligand, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Sarma, P, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-02-06 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
8I9S
 
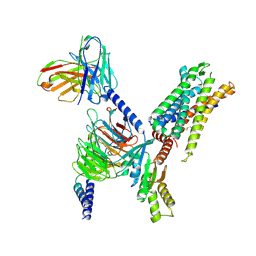 | | Structure of Apo-C3aR-Go complex (Titan) | | Descriptor: | Antibody fragment - ScFv16, C3a anaphylatoxin chemotactic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Sarma, P, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-02-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
8I97
 
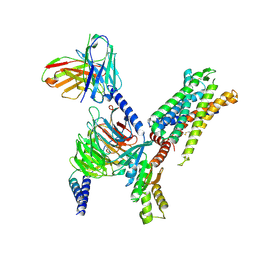 | | Structure of Apo-C3aR-Go complex (Glacios) | | Descriptor: | Antibody fragment - ScFv16, C3a anaphylatoxin chemotactic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Sarma, P, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-02-06 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
8IA2
 
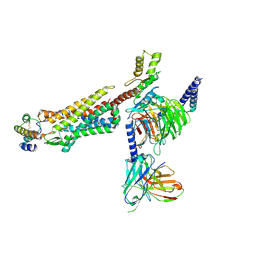 | | Structure of C5a bound human C5aR1 in complex with Go (Composite map) | | Descriptor: | Antibody fragment - ScFv16, C5a anaphylatoxin, C5a anaphylatoxin chemotactic receptor 1, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-02-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
8I9A
 
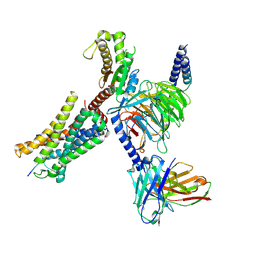 | | Structure of EP54-C3aR-Gq complex | | Descriptor: | Antibody fragment - ScFv16, C3a anaphylatoxin chemotactic receptor, EP54 ligand, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Sarma, P, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-02-06 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
6PGK
 
 | | Membrane Protein Megahertz Crystallography at the European XFEL, Photosystem I XFEL at 2.9 A | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Fromme, R, Gisriel, C, Fromme, P. | | Deposit date: | 2019-06-24 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Membrane protein megahertz crystallography at the European XFEL.
Nat Commun, 10, 2019
|
|
8ILB
 
 | | The complexes of RbcL, AtRaf1 and AtBSD2 (LFB) | | Descriptor: | Protein BUNDLE SHEATH DEFECTIVE 2, chloroplastic, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Wang, R, Song, H, Zhang, W, Wang, N, Zhang, S, Shao, R. | | Deposit date: | 2023-03-03 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the functions of Raf1 and Bsd2 in hexadecameric Rubisco assembly.
Mol Plant, 16, 2023
|
|
8ILM
 
 | | The cryo-EM structure of eight Rubisco large subunits (RbcL), two Arabidopsis thaliana Rubisco accumulation factors 1 (AtRaf1), and seven Arabidopsis thaliana Bundle Sheath Defective 2 (AtBSD2) | | Descriptor: | Protein BUNDLE SHEATH DEFECTIVE 2, chloroplastic, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Wang, R, Song, H, Zhang, W, Wang, N, Zhang, S, Shao, R. | | Deposit date: | 2023-03-03 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the functions of Raf1 and Bsd2 in hexadecameric Rubisco assembly.
Mol Plant, 16, 2023
|
|
