5CF9
 
 | | Cleavage of nicotinamide adenine dinucleotide by the ribosome inactivating protein of Momordica charantia - enzyme-NADP+ co-crystallisation. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NICOTINAMIDE, Ribosome-inactivating protein momordin I | | Authors: | Vinkovic, M, Wood, S.P, Gill, R, Husain, J, Wood, G.E, Dunn, G. | | Deposit date: | 2015-07-08 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Cleavage of nicotinamide adenine dinucleotide by the ribosome-inactivating protein from Momordica charantia.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5NF6
 
 | | Structure of GluK3 ligand-binding domain (S1S2) in complex with CIP-AS at 2.55 A resolution | | Descriptor: | (3~{a}~{S},4~{S},6~{a}~{R})-4,5,6,6~{a}-tetrahydro-3~{a}~{H}-pyrrolo[3,4-d][1,2]oxazole-3,4-dicarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Frydenvang, K, Venskutonyte, R, Thorsen, T.S, Kastrup, J.S. | | Deposit date: | 2017-03-13 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and Affinity of Two Bicyclic Glutamate Analogues at AMPA and Kainate Receptors.
ACS Chem Neurosci, 8, 2017
|
|
5NFF
 
 | | Crystal structure of GP1 receptor binding domain from Morogoro virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, ... | | Authors: | Israeli, H, Cohen-Dvashi, H, Shulman, A, Shimon, A, Diskin, R. | | Deposit date: | 2017-03-14 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.615 Å) | | Cite: | Mapping of the Lassa virus LAMP1 binding site reveals unique determinants not shared by other old world arenaviruses.
PLoS Pathog., 13, 2017
|
|
5NJX
 
 | | Human FKBP51 protein in complex with C-terminal peptide of Human HSP 90-alpha | | Descriptor: | HSP90AA1 protein, Peptidyl-prolyl cis-trans isomerase FKBP5, SULFATE ION | | Authors: | Kumar, R, Moche, M, Winblad, B, Pavlov, P. | | Deposit date: | 2017-03-30 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Combined x-ray crystallography and computational modeling approach to investigate the Hsp90 C-terminal peptide binding to FKBP51.
Sci Rep, 7, 2017
|
|
3R0K
 
 | | Crystal structure of NYSGRC enolase target 200555, a putative dipeptide epimerase from Francisella philomiragia : Tartrate bound, no Mg | | Descriptor: | D(-)-TARTARIC ACID, Enzyme of enolase superfamily, GLYCEROL, ... | | Authors: | Vetting, M.W, Hillerich, B, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-03-08 | | Release date: | 2011-03-30 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5CDL
 
 | | Proline dipeptidase from Deinococcus radiodurans (selenomethionine derivative) | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Proline dipeptidase | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R. | | Deposit date: | 2015-07-04 | | Release date: | 2016-08-10 | | Last modified: | 2018-07-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proline dipeptidase from Deinococcus radiodurans (selenomethionine derivative)
To Be Published
|
|
5LWF
 
 | | Structure of a single domain camelid antibody fragment cAb-G10S in complex with the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | ACETATE ION, Beta-lactamase, Camelid heavy-chain antibody variable fragment cAb-G10S | | Authors: | Vettore, N, Kerff, F, Pain, C, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2016-09-16 | | Release date: | 2017-11-15 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To Be Published
|
|
5CE1
 
 | | Crystal Structure of Serine protease Hepsin in complex with Inhibitor | | Descriptor: | 2-[6-(1-hydroxycyclohexyl)pyridin-2-yl]-1H-indole-5-carboximidamide, Serine protease hepsin | | Authors: | Rao, K.N, Anita, R.C, Sangeetha, R, Anirudha, L, Subramnay, H. | | Deposit date: | 2015-07-06 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Serine protease Hepsin in complex with Inhibitor
To Be Published
|
|
5CMU
 
 | |
2JDW
 
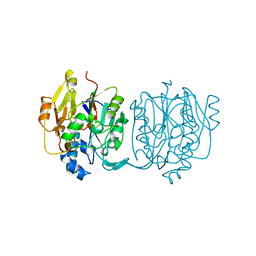 | |
5LYJ
 
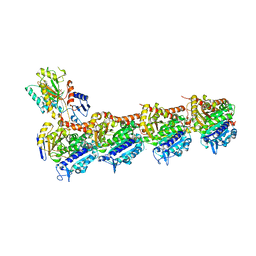 | | Tubulin-Combretastatin A4 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Combretastatin A4, ... | | Authors: | Gaspari, R, Prota, A.E, Cavalli, A, Steinmetz, M.O. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Structural basis of cis- and trans-combretastatin binding to tubulin
Chem, 2017
|
|
3RAI
 
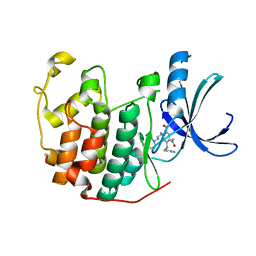 | | CDK2 in complex with inhibitor KVR-1-160 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[3-(morpholin-4-yl)propyl]amino}-5-nitro-2-[(pyridin-3-ylmethyl)amino]benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-28 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
4RBS
 
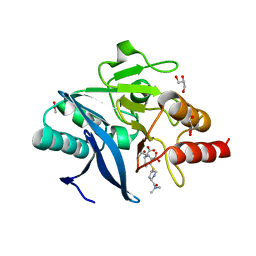 | | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 in the Complex with Hydrolyzed Meropenem | | Descriptor: | (2S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-3-methyl-2H-pyrro le-5-carboxylic acid, ACETIC ACID, Beta-lactamase NDM-1, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, G, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2014-09-12 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 in the Complex with Hydrolyzed Meropenem
To be Published
|
|
5CCQ
 
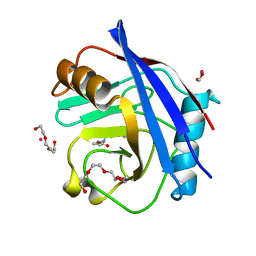 | | Human Cyclophilin D Complexed with Inhibitor | | Descriptor: | FORMIC ACID, POTASSIUM ION, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Gibson, R.P, Shore, E, Kershaw, N, Awais, M, Javed, A, Latawiec, D, Pandalaneni, S, Wen, L, Berry, N, O'Neill, P, Sutton, R, Lian, L.Y. | | Deposit date: | 2015-07-02 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Cyclophilin D Complexed with Inhibitor
To Be Published
|
|
5M51
 
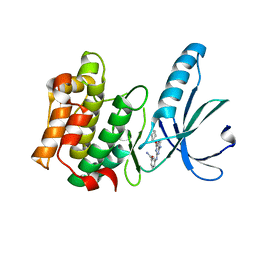 | | Nek2 bound to arylaminopurine compound 8 | | Descriptor: | 3-[[6-(cyclohexylmethoxy)-9~{H}-purin-2-yl]amino]benzamide, Serine/threonine-protein kinase Nek2 | | Authors: | Bayliss, R, Yeoh, S. | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structure-guided design of purine-based probes for selective Nek2 inhibition.
Oncotarget, 8, 2017
|
|
5M53
 
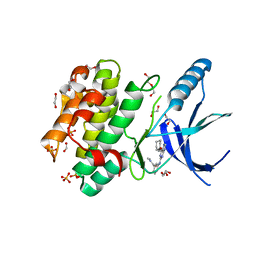 | | Nek2 bound to arylaminopurine inhibitor 11 | | Descriptor: | 1,2-ETHANEDIOL, 3-[[6-(cyclohexylmethoxy)-7~{H}-purin-2-yl]amino]-~{N},~{N}-dimethyl-benzamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bayliss, R, Carr, K.H. | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided design of purine-based probes for selective Nek2 inhibition.
Oncotarget, 8, 2017
|
|
7JQM
 
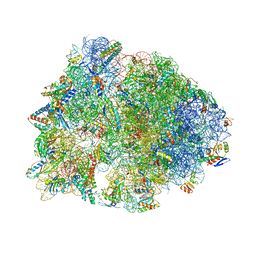 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Bac7-002, mRNA, and deacylated P-site tRNA at 3.05A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mardirossian, M, Sola, R, Beckert, B, Valencic, E, Collis, D.W.P, Borisek, J, Armas, F, Di Stasi, A, Buchmann, J, Syroegin, E.A, Polikanov, Y.S, Magistrato, A, Hilpert, K, Wilson, D.N, Scocchi, M. | | Deposit date: | 2020-08-11 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Peptide Inhibitors of Bacterial Protein Synthesis with Broad Spectrum and SbmA-Independent Bactericidal Activity against Clinical Pathogens.
J.Med.Chem., 63, 2020
|
|
5CD9
 
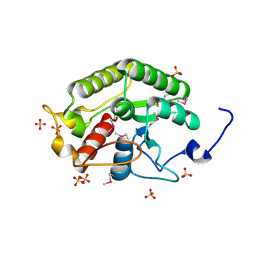 | | Crystal structure of the CTD of Drosophila Oskar protein | | Descriptor: | Maternal effect protein oskar, SULFATE ION | | Authors: | Yang, N, Hu, M, Yu, Z, Wang, M, Lehmann, R, Xu, R.M. | | Deposit date: | 2015-07-03 | | Release date: | 2015-09-02 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structure of Drosophila Oskar reveals a novel RNA binding protein
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2EMT
 
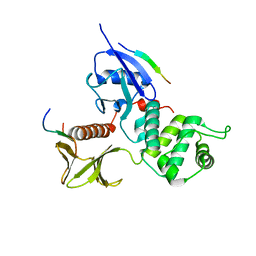 | | Crystal Structure Analysis of the radixin FERM domain complexed with adhesion molecule PSGL-1 | | Descriptor: | P-selectin glycoprotein ligand 1, Radixin | | Authors: | Takai, Y, Kitano, K, Terawaki, S, Maesaki, R, Hakoshima, T. | | Deposit date: | 2007-03-28 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of PSGL-1 binding to ERM proteins
Genes Cells, 12, 2007
|
|
5CDV
 
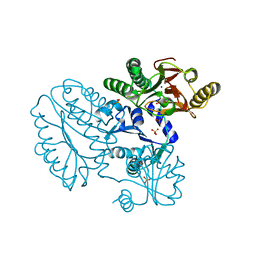 | | Proline dipeptidase from Deinococcus radiodurans R1 | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Proline dipeptidase, ... | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R. | | Deposit date: | 2015-07-05 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Proline dipeptidase from Deinococcus radiodurans R1 at 1.45 Angstrom resolution
To Be Published
|
|
5MKV
 
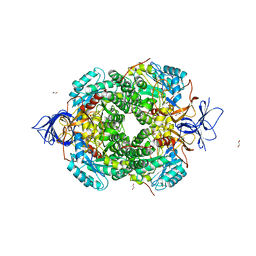 | | Crystal Structure of Human Dihydropyrimidinease-like 2 (DPYSL2A)/Collapsin Response Mediator Protein (CRMP2) residues 13-516 | | Descriptor: | 1,2-ETHANEDIOL, Dihydropyrimidinase-related protein 2 | | Authors: | Sethi, R, Zheng, Y, Krojer, T, Velupillai, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Ahmed, A.A, von Delft, F. | | Deposit date: | 2016-12-05 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tuning microtubule dynamics to enhance cancer therapy by modulating FER-mediated CRMP2 phosphorylation.
Nat Commun, 9, 2018
|
|
2ENR
 
 | | CO-CRYSTALS OF DEMETALLIZED CONCANAVALIN A WITH CADMIUM HAVING A CADMIUM ION BOUND IN BOTH THE S1 SITE AND THE S2 SITE | | Descriptor: | CADMIUM ION, CONCANAVALIN A | | Authors: | Bouckaert, J, Loris, R, Wyns, L. | | Deposit date: | 1998-07-14 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Zinc/calcium- and cadmium/cadmium-substituted concanavalin A: interplay of metal binding, pH and molecular packing.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
7K00
 
 | | Structure of the Bacterial Ribosome at 2 Angstrom Resolution | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Watson, Z.L, Ward, F.R, Meheust, R, Ad, O, Schepartz, A, Banfield, J.F, Cate, J.H.D. | | Deposit date: | 2020-09-02 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (1.98 Å) | | Cite: | Structure of the bacterial ribosome at 2 angstrom resolution.
Elife, 9, 2020
|
|
5MP5
 
 | |
3RM6
 
 | | CDK2 in complex with inhibitor KVR-2-80 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(2-aminopyrimidin-5-yl)methyl]amino}-5-nitro-4-{[2-(piperazin-1-yl)ethyl]amino}benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-04-20 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
