6KMW
 
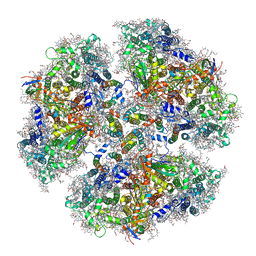 | | Structure of PSI from H. hongdechloris grown under white light condition | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nagao, R, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-08-01 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structural basis for the adaptation and function of chlorophyll f in photosystem I.
Nat Commun, 11, 2020
|
|
6KLC
 
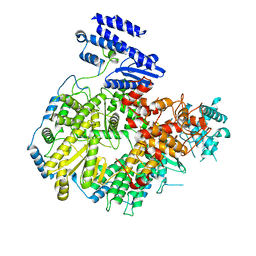 | | Structure of apo Lassa virus polymerase | | Descriptor: | MANGANESE (II) ION, RNA-directed RNA polymerase L | | Authors: | Peng, R, Xu, X, Jing, J, Peng, Q, Gao, G.F, Shi, Y. | | Deposit date: | 2019-07-30 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insight into arenavirus replication machinery.
Nature, 579, 2020
|
|
6QBH
 
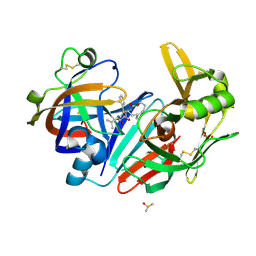 | | Crystal structure of human cathepsin D in complex with macrocyclic inhibitor 33 | | Descriptor: | (4~{S},5~{S},9~{S})-5-oxidanyl-4-(phenylmethyl)-9-propan-2-yl-1-oxa-3,8,11-triazacyclodocosane-2,7,10-trione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brynda, J, Houstecka, R, Majer, P, Mares, M. | | Deposit date: | 2018-12-21 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biomimetic Macrocyclic Inhibitors of Human Cathepsin D: Structure-Activity Relationship and Binding Mode Analysis.
J.Med.Chem., 63, 2020
|
|
6QEY
 
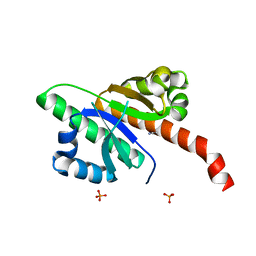 | | IMP1 KH1 and KH2 domains create a structural platform with unique RNA recognition and re-modelling properties | | Descriptor: | ACETONITRILE, Insulin-like growth factor 2 mRNA-binding protein 1, PHOSPHATE ION | | Authors: | Dagil, R, Ball, N.J, Ogrodowicz, R.W, Purkiss, A.G, Taylor, I.A, Ramos, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | IMP1 KH1 and KH2 domains create a structural platform with unique RNA recognition and re-modelling properties.
Nucleic Acids Res., 47, 2019
|
|
6QBU
 
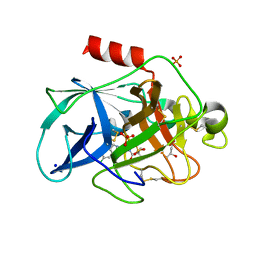 | | Crystal structure of Porcine Pancreatic Elastase (PPE) in complex with the 3-Oxo-beta-Sultam inhibitor LMC188 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Chymotrypsin-like elastase family member 1, PHOSPHATE ION, ... | | Authors: | Brito, J.A, Almeida, V.T, Carvalho, L.M, Moreira, R, Archer, M. | | Deposit date: | 2018-12-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | 3-Oxo-beta-sultam as a Sulfonylating Chemotype for Inhibition of Serine Hydrolases and Activity-Based Protein Profiling.
Acs Chem.Biol., 15, 2020
|
|
6JT7
 
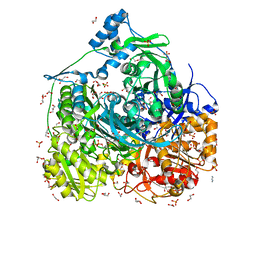 | | Crystal structure of 452-453_deletion mutant of FGAM Synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Sharma, N, Ahalawat, N, Sandhu, P, Mondal, J, Anand, R. | | Deposit date: | 2019-04-10 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Role of allosteric switches and adaptor domains in long-distance cross-talk and transient tunnel formation.
Sci Adv, 6, 2020
|
|
4U8I
 
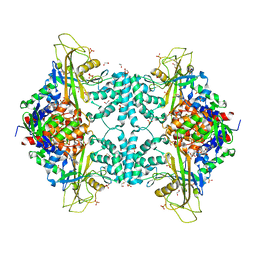 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant F66A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
6QJ5
 
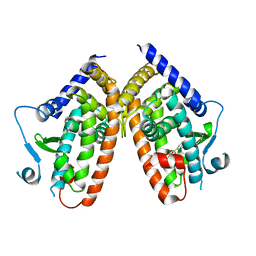 | | X-ray structure of PPARgamma LBD with the ligand NV1380 | | Descriptor: | (2~{S})-3-methyl-2-[(4-octoxyphenyl)carbonylamino]butanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Capelli, D. | | Deposit date: | 2019-01-22 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Novel N-Substituted Valine Derivative with Unique Peroxisome Proliferator-Activated Receptor gamma Binding Properties and Biological Activities.
J.Med.Chem., 63, 2020
|
|
6K0V
 
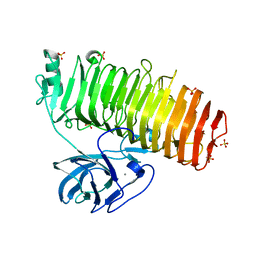 | | Catalytic domain of GH87 alpha-1,3-glucanase D1069A in complex with tetrasaccharides | | Descriptor: | Alpha-1,3-glucanase, CALCIUM ION, SULFATE ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6KB9
 
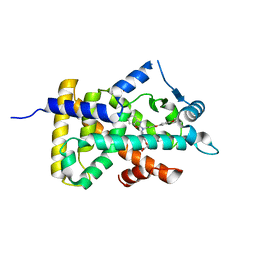 | | X-ray structure of human PPARalpha ligand binding domain-pemafibrate co-crystals obtained by cross-seeding | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6QCB
 
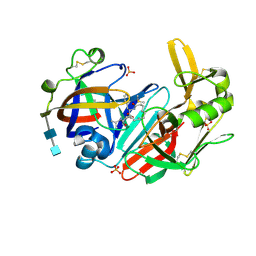 | | Crystal structure of human cathepsin D in complex with macrocyclic inhibitor 9 | | Descriptor: | (3~{S},7~{S},8~{S})-7-oxidanyl-8-(phenylmethyl)-3-propan-2-yl-1,4,9-triazacyclohenicosane-2,5,10-trione, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cathepsin D, ... | | Authors: | Brynda, J, Houstecka, R, Majer, P, Mares, M. | | Deposit date: | 2018-12-27 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biomimetic Macrocyclic Inhibitors of Human Cathepsin D: Structure-Activity Relationship and Binding Mode Analysis.
J.Med.Chem., 63, 2020
|
|
6KJL
 
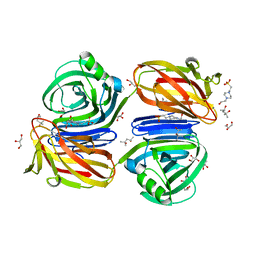 | | Xylanase J from Bacillus sp. strain 41M-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Manami, S, Teisuke, T, Nakatani, K, Katano, K, Kojima, K, Saka, N, Mikami, B, Yatsunami, R, Nakamura, S, Yasukawa, K. | | Deposit date: | 2019-07-22 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Increase in the thermostability of GH11 xylanase XynJ from Bacillus sp. strain 41M-1 using site saturation mutagenesis.
Enzyme.Microb.Technol., 130, 2019
|
|
6KLD
 
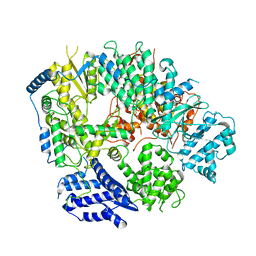 | | Structure of apo Machupo virus polymerase | | Descriptor: | MANGANESE (II) ION, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Peng, R, Xu, X, Jing, J, Peng, Q, Gao, G.F, Shi, Y. | | Deposit date: | 2019-07-30 | | Release date: | 2020-03-18 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural insight into arenavirus replication machinery.
Nature, 579, 2020
|
|
6QIQ
 
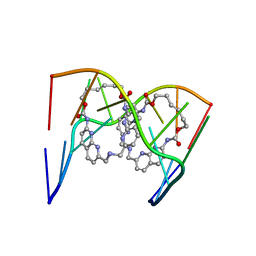 | | Crystal structure of seleno-derivative CAG repeats with synthetic CMBL3a compound | | Descriptor: | CMBL3a, RNA (5'-R(*GP*CP*AP*G)-D(P*(CSL))-R(P*AP*GP*C)-3') | | Authors: | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Micura, R, Nakatani, K. | | Deposit date: | 2019-01-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.519 Å) | | Cite: | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
6QLC
 
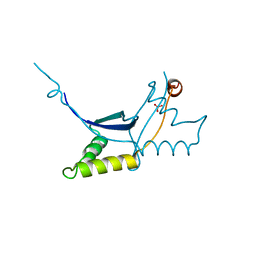 | | The ssDNA-binding RNA polymerase cofactor Drc from Pseudomonas phage LUZ7 | | Descriptor: | PHOSPHATE ION, ssDNA binding RNA Polymerase cofactor | | Authors: | De Zitter, E, Boon, M, De Smet, J, Lavigne, R, Van Meervelt, L. | | Deposit date: | 2019-01-31 | | Release date: | 2019-10-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 'Drc', a structurally novel ssDNA-binding transcription regulator of N4-related bacterial viruses.
Nucleic Acids Res., 48, 2020
|
|
6Q52
 
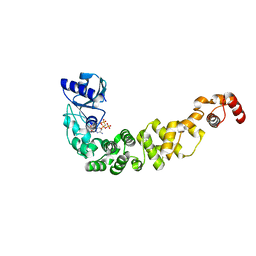 | | Structure of a psychrophilic CCA-adding enzyme in complex with CMPcPP at room temperature in ChipX microfluidic device | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, CCA-adding enzyme | | Authors: | de Wijn, R, Hennig, O, Rollet, K, Bluhm, A, Betat, H, Moerl, M, Lorber, B, Sauter, C. | | Deposit date: | 2018-12-06 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
6JT8
 
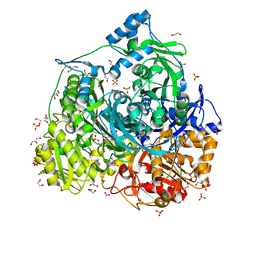 | | Crystal structure of 450-451_deletion mutant of FGAM Synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Sharma, N, Ahalawat, N, Sandhu, P, Mondal, J, Anand, R. | | Deposit date: | 2019-04-10 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of allosteric switches and adaptor domains in long-distance cross-talk and transient tunnel formation.
Sci Adv, 6, 2020
|
|
4U8P
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant Y317A complexed with UDP | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
6K0S
 
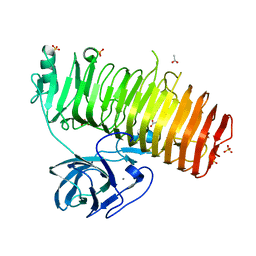 | | Catalytic domain of GH87 alpha-1,3-glucanase D1069A in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6KB5
 
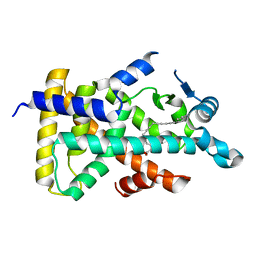 | | X-ray structure of human PPARalpha ligand binding domain-5,8,11,14-eicosatetraynoic Acid (ETYA) co-crystals obtained by delipidation and cross-seeding | | Descriptor: | GLYCEROL, Peroxisome proliferator-activated receptor alpha, icosa-5,8,11,14-tetraynoic acid | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6QID
 
 | | Crystal structure of DEAH-box ATPase Prp43-S387A | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Hamann, F, Ficner, R, Enders, M. | | Deposit date: | 2019-01-18 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural basis for RNA translocation by DEAH-box ATPases.
Nucleic Acids Res., 47, 2019
|
|
6KFZ
 
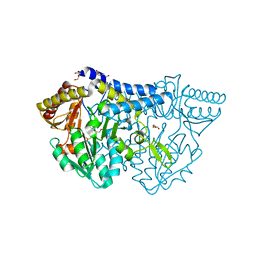 | | SufS from Bacillus subtilis, soaked with L-cysteine for 90 sec at 1.96 angstrom resolution | | Descriptor: | Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-CYSTEINE | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2019-07-09 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Snapshots of PLP-substrate and PLP-product external aldimines as intermediates in two types of cysteine desulfurase enzymes.
Febs J., 287, 2020
|
|
6QFI
 
 | | Structure of human Mcl-1 in complex with BIM BH3 peptide | | Descriptor: | Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6QFQ
 
 | | Structure of human Mcl-1 in complex with indole acid inhibitor | | Descriptor: | 7-(3,5-dimethyl-1~{H}-pyrazol-4-yl)-3-(3-naphthalen-1-yloxypropyl)-1~{H}-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6QG8
 
 | | Structure of human Bcl-2 in complex with PUMA BH3 peptide | | Descriptor: | Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Bcl-2-like protein 1, Bcl-2-binding component 3 | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
