4WNX
 
 | | Netrin 4 lacking the C-terminal Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | McDougall, M, Patel, T, Reuten, R, Meier, M, Koch, M, Stetefeld, J. | | Deposit date: | 2014-10-14 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.723 Å) | | Cite: | Structural decoding of netrin-4 reveals a regulatory function towards mature basement membranes.
Nat Commun, 7, 2016
|
|
4WOY
 
 | | Crystal structure and functional analysis of MiD49, a receptor for the mitochondrial fission protein Drp1 | | Descriptor: | Mitochondrial dynamics protein MID49 | | Authors: | Loson, O.C, Meng, S, Ngo, H.B, Liu, R, Kaiser, J.T, Chan, D.C. | | Deposit date: | 2014-10-17 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and functional analysis of MiD49, a receptor for the mitochondrial fission protein Drp1.
Protein Sci., 24, 2015
|
|
1GZX
 
 | | Oxy T State Haemoglobin - Oxygen bound at all four haems | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, OXYGEN MOLECULE, ... | | Authors: | Paoli, M, Liddington, R, Tame, J, Wilkinson, A, Dodson, G. | | Deposit date: | 2002-06-07 | | Release date: | 2002-07-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of T State Haemoglobin with Oxygen Bound at All Four Haems.
J.Mol.Biol., 256, 1996
|
|
4WUV
 
 | | Crystal Structure of a putative D-Mannonate oxidoreductase from Haemophilus influenza (Avi_5165, TARGET EFI-513796) with bound NAD | | Descriptor: | 1,2-ETHANEDIOL, 2-hydroxycyclohexanecarboxyl-CoA dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2014-11-03 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Crystal structure of a putative D-Mannonate oxidoreductase from Haemophilus influenza (Avi_5165, TARGET EFI-513796) with bound NAD
To be published
|
|
1HA8
 
 | |
4WXE
 
 | | CRYSTAL STRUCTURE OF A LACI REGULATOR FROM LACTOBACILLUS CASEI (LSEI_2103, TARGET EFI-512911) WITH BOUND TRIS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-13 | | Release date: | 2014-11-26 | | Last modified: | 2015-10-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF A LACI REGULATOR FROM LACTOBACILLUS CASEI (LSEI_2103, TARGET EFI-512911) WITH BOUND TRIS
To be published
|
|
1H7O
 
 | | SCHIFF-BASE COMPLEX OF YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE WITH 5-AMINOLAEVULINIC ACID AT 1.7 A RESOLUTION | | Descriptor: | 5-AMINOLAEVULINIC ACID DEHYDRATASE, DELTA-AMINO VALERIC ACID, ZINC ION | | Authors: | Erskine, P.T, Newbold, R, Brindley, A.A, Wood, S.P, Shoolingin-Jordan, P.M, Warren, M.J, Cooper, J.B. | | Deposit date: | 2001-07-09 | | Release date: | 2001-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Substrate and Three Inhibitors
J.Mol.Biol., 312, 2001
|
|
4X8F
 
 | | Vibrio cholerae O395 Ribokinase in apo form | | Descriptor: | Ribokinase | | Authors: | Paul, R, Patra, M.D, Sen, U. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of Apo and Ligand
Bound Vibrio choleraeRibokinase (Vc-RK):
Role of Monovalent Cation Induced Activation
and Structural Flexibility in Sugar
Phosphorylation
Adv Exp Med Biol., 842, 2015
|
|
1H8F
 
 | | Glycogen Synthase Kinase 3 beta. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCOGEN SYNTHASE KINASE-3 BETA | | Authors: | Dajani, R, Pearl, L.H, Roe, S.M. | | Deposit date: | 2001-02-05 | | Release date: | 2002-01-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Glycogen Synthase Kinase 3Beta . Structural Basis for Phosphate-Primed Substrate Specificity and Autoinhibition
Cell(Cambridge,Mass.), 105, 2001
|
|
7B9E
 
 | | Crystal structure of MurE from E.coli in complex with Z275151340 | | Descriptor: | 4-chloro-N-cyclopentyl-1-methyl-1H-pyrazole-3-carboxamide, ISOPROPYL ALCOHOL, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-14 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
1GU8
 
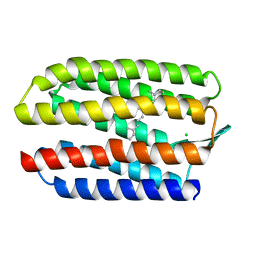 | | SENSORY RHODOPSIN II | | Descriptor: | CHLORIDE ION, RETINAL, SENSORY RHODOPSIN II | | Authors: | Edman, K, Royant, A, Nollert, P, Maxwell, C.A, Pebay-Peyroula, E, Navarro, J, Neutze, R, Landau, E.M. | | Deposit date: | 2002-01-24 | | Release date: | 2002-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Early Structural Rearrangements in the Photocycle of an Integral Membrane Sensory Receptor
Structure, 10, 2002
|
|
7BQ3
 
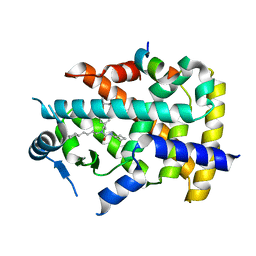 | | X-ray structure of human PPARalpha ligand binding domain-GW7647-SRC1 coactivator peptide co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-03-23 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
5KE4
 
 | | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) containing loop C from the human alpha 6 nicotinic acetylcholine receptor in complex with 2-((5-(3,7-Diazabicyclo[3.3.1]nonan-3-yl)pyridin-3-yl)oxy)- N,N-dimethylethanamine (BPC) | | Descriptor: | 2-((5-(3,7-Diazabicyclo[3.3.1]nonan-3-yl)pyridin-3-yl)oxy)-N,N-dimethylethanamine, Soluble acetylcholine receptor | | Authors: | Bobango, J, Wu, J, Talley, I.T, Ralston, R, Sankaran, B, Talley, T.T. | | Deposit date: | 2016-06-09 | | Release date: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) containing loop C from the human alpha 6 nicotinic acetylcholine receptor in complex with 2-((5-(3,7-Diazabicyclo[3.3.1]nonan-3-yl)pyridin-3-yl)oxy)-
N,N-dimethylethanamine (BPC)
To Be Published
|
|
4Z9X
 
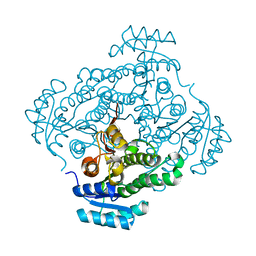 | | Crystal structure of 2-keto-3-deoxy-D-gluconate dehydrogenase from Streptococcus pyogenes | | Descriptor: | Gluconate 5-dehydrogenase | | Authors: | Maruyama, Y, Takase, R, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-04-13 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural determinants in bacterial 2-keto-3-deoxy-D-gluconate dehydrogenase KduD for dual-coenzyme specificity
Proteins, 84, 2016
|
|
4YYR
 
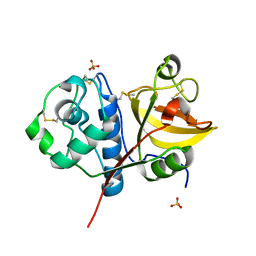 | | Ficin B crystal form I | | Descriptor: | Ficin B, SULFATE ION | | Authors: | Azarkan, M, Baeyens-Volant, D, Loris, R. | | Deposit date: | 2015-03-24 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Crystal structure of four variants of the protease Ficin from the common fig
To Be Published
|
|
7BS8
 
 | | Bovine Pancreatic Trypsin with Benzamidine (Cryo) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Maeki, M, Ito, S, Takeda, R, Funakubo, T, Ueno, G, Ishida, A, Tani, H, Yamamoto, M, Tokeshi, M. | | Deposit date: | 2020-03-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Room-temperature crystallography using a microfluidic protein crystal array device and its application to protein-ligand complex structure analysis.
Chem Sci, 11, 2020
|
|
3G9Q
 
 | | Crystal structure of the FhuD fold-family BSU3320, a periplasmic binding protein component of a Fep/Fec-like ferrichrome ABC transporter from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR577A | | Descriptor: | Ferrichrome-binding protein | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Janjua, H, Mao, L, Xiao, R, Ciccosanti, C, Foote, E.L, Lee, D, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-13 | | Release date: | 2009-03-03 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the FhuD fold-family BSU3320, a periplasmic binding protein component of a Fep/Fec-like ferrichrome ABC transporter from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR577A.
To be Published
|
|
4ZLX
 
 | |
1LAU
 
 | | URACIL-DNA GLYCOSYLASE | | Descriptor: | DNA (5'-D(*TP*TP*T)-3'), PROTEIN (URACIL-DNA GLYCOSYLASE (E.C.3.2.2.-)) | | Authors: | Pearl, L.H, Savva, R. | | Deposit date: | 1996-01-03 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of specific base-excision repair by uracil-DNA glycosylase.
Nature, 373, 1995
|
|
8R1O
 
 | | Structure of C. thermophilum RNA exosome core | | Descriptor: | Exoribonuclease phosphorolytic domain-containing protein, Exoribonuclease-like protein, Exosome complex component MTR3, ... | | Authors: | Lazzaretti, D, Liebau, J, Pilsl, M, Sprangers, R. | | Deposit date: | 2023-11-02 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Beyond static structures: quantitative dynamics in the eukaryotic RNA exosome complex
Biorxiv, 2024
|
|
7BY1
 
 | |
7BZT
 
 | | Cryo-EM structure of mature Coxsackievirus A10 in complex with KRM1 at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2020-08-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4Z32
 
 | |
8R2J
 
 | |
1OWL
 
 | | Structure of apophotolyase from Anacystis nidulans | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
