6PBX
 
 | |
4IGF
 
 | | Crystal structure of Plasmodium falciparum FabI complexed with NAD and inhibitor 3-(4-Chloro-2-hydroxyphenoxy)-7-hydroxy-2H-chromen-2-one | | Descriptor: | 3-(4-chloro-2-hydroxyphenoxy)-7-hydroxy-2H-chromen-2-one, Enoyl-acyl carrier reductase, GLYCEROL, ... | | Authors: | Kostrewa, D, Perozzo, R. | | Deposit date: | 2012-12-17 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, Synthesis, and Biological and Crystallographic Evaluation of Novel Inhibitors of Plasmodium falciparum Enoyl-ACP-reductase (PfFabI)
J.Med.Chem., 56, 2013
|
|
1FGD
 
 | |
7QNG
 
 | | Structure of a MHC I-Tapasin-ERp57 complex | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Mueller, I.K, Thomas, C, Trowitzsch, S, Tampe, R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of an MHC I-tapasin-ERp57 editing complex defines chaperone promiscuity.
Nat Commun, 13, 2022
|
|
4Y6M
 
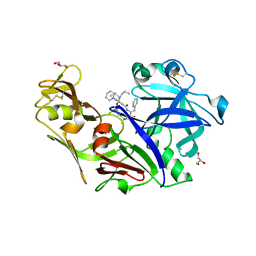 | | Structure of plasmepsin II from Plasmodium falciparum complexed with inhibitor PG418 | | Descriptor: | GLYCEROL, Plasmepsin-2, ~{N}1-[(~{Z},3~{R})-4-[2-(3-methoxyphenyl)propan-2-ylamino]-3-oxidanyl-1-phenyl-but-1-en-2-yl]-5-piperidin-1-yl-~{N}3,~{N}3-dipropyl-benzene-1,3-dicarboxamide | | Authors: | Recacha, R, Akopjana, I, Tars, K, Jaudzems, K. | | Deposit date: | 2015-02-13 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structures of plasmepsin II from Plasmodium falciparum in complex with two hydroxyethylamine-based inhibitors.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
7TZD
 
 | |
4YP2
 
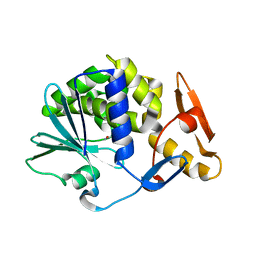 | | Cleavage of nicotinamide adenine dinucleotides by the ribosome inactivating protein from Momordica charantia | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NICOTINAMIDE, Ribosome-inactivating protein momordin I | | Authors: | Vinkovic, M, Hussain, J, Wood, G.E, Gill, R, Wood, S.P. | | Deposit date: | 2015-03-12 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Cleavage of nicotinamide adenine dinucleotide by the ribosome-inactivating protein from Momordica charantia.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
7TZ8
 
 | |
7Q9E
 
 | | CYP106A1 | | Descriptor: | COBALT (II) ION, Cytochrome P450, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Carius, Y, Hutter, M, Kiss, F, Bernhardt, R, Lancaster, C.R.D. | | Deposit date: | 2021-11-12 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural comparison of the cytochrome P450 enzymes CYP106A1 and CYP106A2 provides insight into their differences in steroid conversion.
Febs Lett., 596, 2022
|
|
7QPD
 
 | | Structure of the human MHC I peptide-loading complex editing module | | Descriptor: | Beta-2-microglobulin, Calreticulin, HLA class I histocompatibility antigen, ... | | Authors: | Domnick, A, Susac, L, Trowitzsch, S, Thomas, C, Tampe, R. | | Deposit date: | 2022-01-03 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Molecular basis of MHC I quality control in the peptide loading complex.
Nat Commun, 13, 2022
|
|
4YS6
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM CLOSTRIDIUM PHYTOFERMENTANS (Cphy_1585, TARGET EFI-511156) WITH BOUND BETA-D-GLUCOSE | | Descriptor: | CHLORIDE ION, Putative solute-binding component of ABC transporter, SODIUM ION, ... | | Authors: | Vetting, M.W, Patskovsky, Y, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-16 | | Release date: | 2015-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM CLOSTRIDIUM PHYTOFERMENTANS (Cphy_1585, TARGET EFI-511156) WITH BOUND BETA-D-GLUCOSE
To be published
|
|
1FGN
 
 | | MONOCLONAL MURINE ANTIBODY 5G9-ANTI-HUMAN TISSUE FACTOR | | Descriptor: | IMMUNOGLOBULIN FAB 5G9 | | Authors: | Huang, M, Syed, R, Stura, E.A, Stone, M.J, Stefanko, R.S, Ruf, W, Edgington, T.S, Wilson, I.A. | | Deposit date: | 1997-04-10 | | Release date: | 1998-02-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The mechanism of an inhibitory antibody on TF-initiated blood coagulation revealed by the crystal structures of human tissue factor, Fab 5G9 and TF.5G9 complex.
J.Mol.Biol., 275, 1998
|
|
8E2O
 
 | |
8ED9
 
 | | cryo-EM structure of TRPM3 ion channel in the presence with PIP2 and PregS, state 2 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-09-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
1FDT
 
 | | HUMAN 17-BETA-HYDROXYSTEROID-DEHYDROGENASE TYPE 1 COMPLEXED WITH ESTRADIOL AND NADP+ | | Descriptor: | 17-BETA-HYDROXYSTEROID-DEHYDROGENASE, ESTRADIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Housset, D, Breton, R, Mazza, C, Fontecilla-Camps, J.-C. | | Deposit date: | 1996-06-28 | | Release date: | 1997-02-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a complex of human 17beta-hydroxysteroid dehydrogenase with estradiol and NADP+ identifies two principal targets for the design of inhibitors.
Structure, 4, 1996
|
|
6PJD
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-32 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,4S,5S)-4-hydroxy-5-{[N-(methoxycarbonyl)-3-methyl-L-valyl]amino}-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
6PJM
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-35 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3S,5S)-3-hydroxy-5-{[N-(methoxycarbonyl)-3-methyl-L-valyl]amino}-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
3TPC
 
 | | Crystal structure of a hypothtical protein SMa1452 from Sinorhizobium meliloti 1021 | | Descriptor: | Short chain alcohol dehydrogenase-related dehydrogenase | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-09-07 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of a hypothtical protein SMa1452 from Sinorhizobium meliloti 1021
To be Published
|
|
6PLJ
 
 | | A nucleotidyl transferase from Methanothermobacter thermautotroptrophicus (Mth528) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SODIUM ION, ... | | Authors: | Carbone, V, Schofield, L, Sutherland-Smith, A, Ronimus, R. | | Deposit date: | 2019-07-01 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A nucleotidyl transferase from Methanothermobacter thermautotroptrophicus (Mth528)
To Be Published
|
|
4YVW
 
 | | crystal structure of an enterovirus 71/coxsackievirus A16 chimeric virus-like particle | | Descriptor: | Capsid protein VP0, Capsid protein VP1, Capsid protein VP3 | | Authors: | Chen, R, Lyu, K. | | Deposit date: | 2015-03-20 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal Structures of Yeast-Produced Enterovirus 71 and Enterovirus 71/Coxsackievirus A16 Chimeric Virus-Like Particles Provide the Structural Basis for Novel Vaccine Design against Hand-Foot-and-Mouth Disease
J.Virol., 89, 2015
|
|
1FVN
 
 | |
8SYP
 
 | | Genomic CX3CR1 nucleosome | | Descriptor: | DNA (162-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | Authors: | Lian, T, Guan, R, Bai, Y. | | Deposit date: | 2023-05-25 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural mechanism of synergistic targeting of the CX3CR1 nucleosome by PU.1 and C/EBP alpha.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7Q6P
 
 | |
1G0F
 
 | |
8SO0
 
 | | Cryo-EM structure of the PP2A:B55-FAM122A complex | | Descriptor: | FE (III) ION, PPP2R1A-PPP2R2A-interacting phosphatase regulator 1, Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform, ... | | Authors: | Fuller, J.R, Padi, S.K.R, Peti, W, Page, R. | | Deposit date: | 2023-04-28 | | Release date: | 2023-10-25 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.79961 Å) | | Cite: | Cryo-EM structures of PP2A:B55-FAM122A and PP2A:B55-ARPP19.
Nature, 625, 2024
|
|
