7UOS
 
 | | Structure of WNK1 inhibitor complex | | Descriptor: | 1,2-ETHANEDIOL, 4-[bromo(dichloro)methanesulfonyl]-N-cyclohexyl-2-nitroaniline, Serine/threonine-protein kinase WNK1 | | Authors: | Akella, R, Goldsmith, E.J, Akella, R. | | Deposit date: | 2022-04-13 | | Release date: | 2023-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of WNK1 inhibitor complex
To Be Published
|
|
7XM1
 
 | | Cryo-EM structure of mTIP60-Ba (metal-ion induced TIP60 (K67E) complex with barium ions | | Descriptor: | BARIUM ION, TIP60 K67E mutant | | Authors: | Ohara, N, Kawakami, N, Arai, R, Adachi, N, Moriya, T, Kawasaki, M, Miyamoto, K. | | Deposit date: | 2022-04-24 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Reversible Assembly of an Artificial Protein Nanocage Using Alkaline Earth Metal Ions.
J.Am.Chem.Soc., 145, 2023
|
|
1OJH
 
 | | Crystal structure of NblA from PCC 7120 | | Descriptor: | 1,2-ETHANEDIOL, NBLA | | Authors: | Bienert, R, Baier, K, Lockau, W, Heinemann, U. | | Deposit date: | 2003-07-10 | | Release date: | 2004-07-15 | | Last modified: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Nbla from Anabaena Sp. Pcc 7120, a Small Protein Playing a Key Role in Phycobilisome Degradation.
J.Biol.Chem., 281, 2006
|
|
3RCY
 
 | | CRYSTAL STRUCTURE OF Mandelate racemase/muconate lactonizing enzyme-like protein from Roseovarius sp. TM1035 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme-like protein, ... | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-03-31 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | CRYSTAL STRUCTURE OF Mandelate racemase/muconate lactonizing enzyme-like protein from Roseovarius sp. TM1035
To be Published
|
|
3RQS
 
 | | Crystal Structure of human L-3- Hydroxyacyl-CoA dehydrogenase (EC1.1.1.35) from mitochondria at the resolution 2.0 A, Northeast Structural Genomics Consortium Target HR487, Mitochondrial Protein Partnership | | Descriptor: | GLYCEROL, Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2011-04-28 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Northeast Structural Genomics Consortium Target HR487
To be published
|
|
3RHD
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase GapN from Methanocaldococcus jannaschii DSM 2661 complexed with NADP | | Descriptor: | Lactaldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-04-11 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase GapN from Methanocaldococcus jannaschii DSM 2661 complexed with NADP
To be Published
|
|
1QM3
 
 | |
1QOO
 
 | | lectin UEA-II complexed with NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHITIN BINDING LECTIN, ... | | Authors: | Loris, R, De Greve, H, Dao-Thi, M.-H, Messens, J, Imberty, A, Wyns, L. | | Deposit date: | 1999-11-15 | | Release date: | 1999-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by Lectin II from Ulex Europaeus, a Protein with a Promiscuous Carbohydrate Binding Site
J.Mol.Biol., 301, 2000
|
|
1QFD
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-AMYLASE INHIBITOR (AAI) | | Descriptor: | PROTEIN (ALPHA-AMYLASE INHIBITOR) | | Authors: | Lu, S, Deng, P, Liu, X, Luo, J, Han, R, Gu, X, Liang, S, Wang, X, Feng, L, Lozanov, V, Patthy, A, Pongor, S. | | Deposit date: | 1999-04-08 | | Release date: | 1999-07-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the major alpha-amylase inhibitor of the crop plant amaranth.
J.Biol.Chem., 274, 1999
|
|
3BZ0
 
 | | Lactobacillus Casei Thymidylate Synthase Ternary Complex with DUMP and the Phtalimidic Derivative C00 | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-(4-hydroxybiphenyl-3-yl)-4-methyl-1H-isoindole-1,3(2H)-dione, Thymidylate synthase | | Authors: | Leone, R, Cancian, L, Costi, M.P, Ferrari, S, Luciani, R, Mangani, S. | | Deposit date: | 2008-01-17 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of the binding modes of N-phenylphthalimides inhibiting bacterial thymidylate synthase through X-ray crystallography screening
J.Med.Chem., 54, 2011
|
|
2PL8
 
 | | D(GTATACC) under hydrostatic pressure of 1.04 GPa | | Descriptor: | 5'-D(*DGP*DGP*DTP*DAP*DTP*DAP*DCP*DC)-3', SPERMINE | | Authors: | Prange, T, Girard, E, Fourme, R, Kahn, R. | | Deposit date: | 2007-04-19 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Adaptation of the base-paired double-helix molecular architecture to extreme pressure.
Nucleic Acids Res., 35, 2007
|
|
8TOS
 
 | | ACE2-peptide 6 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Christie, M, Payne, R. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
218D
 
 | | THE STRUCTURE OF A NEW CRYSTAL FORM OF A DNA DODECAMER CONTAINING T.(O6ME)G BASE PAIRS | | Descriptor: | DNA (5'-D(*CP*GP*TP*GP*AP*AP*TP*TP*CP*(6OG)P*CP*G)-3') | | Authors: | Vojtechovsky, J, Eaton, M.D, Gaffney, B, Jones, R, Berman, H.M. | | Deposit date: | 1995-07-18 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of a new crystal form of a DNA dodecamer containing T.(O6Me)G base pairs.
Biochemistry, 34, 1995
|
|
3BNZ
 
 | | Crystal structure of Thymidylate Synthase ternary complex with dUMP and 8A inhibitor | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 4-(4-methyl-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)benzonitrile, PHOSPHATE ION, ... | | Authors: | Leone, R, Cancian, L, Luciani, R, Ferrari, S, Costi, M.P, Mangani, S. | | Deposit date: | 2007-12-15 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of the binding modes of N-phenylphthalimides inhibiting bacterial thymidylate synthase through X-ray crystallography screening
J.Med.Chem., 54, 2011
|
|
3BYX
 
 | | Lactobacillus CASEI Thymidylate Synthase Ternary Complex with DUMP and the Phtalimidic Derivative C00 in Multiple Binding Modes | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-(4-hydroxybiphenyl-3-yl)-4-methyl-1H-isoindole-1,3(2H)-dione, Thymidylate synthase | | Authors: | Leone, R, Mangani, S, Cancian, L, Costi, M.P, Luciani, R, Ferrari, S. | | Deposit date: | 2008-01-16 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of the binding modes of N-phenylphthalimides inhibiting bacterial thymidylate synthase through X-ray crystallography screening
J.Med.Chem., 54, 2011
|
|
3RHH
 
 | | Crystal structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Bacillus halodurans C-125 complexed with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-04-11 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Bacillus halodurans C-125 complexed with NADP
To be Published
|
|
3C06
 
 | | Lactobacillus CASEI Thymidylate Synthase Ternary Complex With DUMP and the Phtalimidic Derivative 14C in Multiple Binding Modes-Mode 1 | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-(2-chloropyridin-4-yl)-4-methyl-1H-isoindole-1,3(2H)-dione, Thymidylate synthase | | Authors: | Leone, R, Mangani, S, Cancian, L, Costi, M.P, Luciani, R, Ferrari, S. | | Deposit date: | 2008-01-18 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of the binding modes of N-phenylphthalimides inhibiting bacterial thymidylate synthase through X-ray crystallography screening
J.Med.Chem., 54, 2011
|
|
1YMA
 
 | |
1QLZ
 
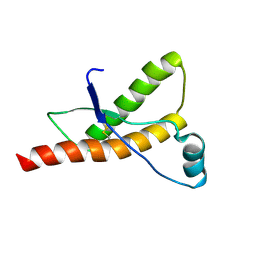 | | Human prion protein | | Descriptor: | PRION PROTEIN | | Authors: | Zahn, R, Liu, A, Luhrs, T, Wuthrich, K. | | Deposit date: | 1999-09-20 | | Release date: | 1999-12-16 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Human Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2ZIE
 
 | | Crystal Structure of TTHA0409, Putatative DNA Modification Methylase from Thermus thermophilus HB8- Selenomethionine derivative | | Descriptor: | Putative modification methylase | | Authors: | Morita, R, Ishikawa, H, Nakagawa, N, Masui, R, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-02-15 | | Release date: | 2008-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative DNA methylase TTHA0409 from Thermus thermophilus HB8
Proteins, 73, 2008
|
|
2KD0
 
 | | NMR solution structure of O64736 protein from Arabidopsis thaliana. Northeast Structural Genomics Consortium MEGA Target AR3445A | | Descriptor: | LRR repeats and ubiquitin-like domain-containing protein At2g30105 | | Authors: | Swapna, G.V.T, Shastry, R, Foote, E, Ciccosanti, C, Jiang, M, Xiao, R, Nair, R, Everett, J, Huang, Y, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-31 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of O64736 protein from Arabidopsis thaliana
To be Published
|
|
2KRK
 
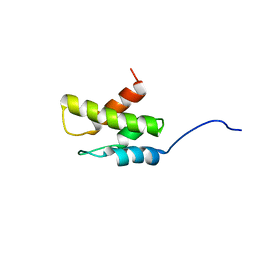 | | Solution NMR Structure of 26S protease regulatory subunit 8 from H.sapiens, Northeast Structural Genomics Consortium Target Target HR3102A | | Descriptor: | 26S protease regulatory subunit 8 | | Authors: | Liu, G, Janjua, J, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-12 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of 26S protease regulatory subunit 8 from H.sapiens, Northeast Structural Genomics Consortium Target HR3102A
To be Published
|
|
3CAN
 
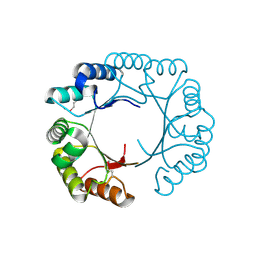 | | Crystal structure of a domain of pyruvate-formate lyase-activating enzyme from Bacteroides vulgatus ATCC 8482 | | Descriptor: | Pyruvate-formate lyase-activating enzyme | | Authors: | Nocek, B, Hendricks, R, Hatzos, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-20 | | Release date: | 2008-03-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a domain of pyruvate-formate lyase-activating enzyme from Bacteroides vulgatus ATCC 8482.
To be Published
|
|
1QNW
 
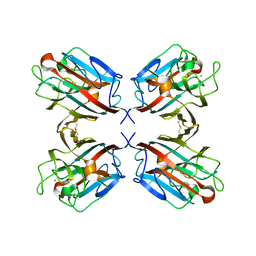 | | lectin II from Ulex europaeus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHITIN BINDING LECTIN, ... | | Authors: | Loris, R, De Greve, H, Dao-Thi, M.-H, Messens, J, Imberty, A, Wyns, L. | | Deposit date: | 1999-10-22 | | Release date: | 1999-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by Lectin II from Ulex Europaeus, a Protein with a Promiscuous Carbohydrate Binding Site
J.Mol.Biol., 301, 2000
|
|
1QT9
 
 | | OXIDIZED [2FE-2S] FERREDOXIN FROM ANABAENA PCC7119 | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN I | | Authors: | Morales, R, Chron, M.-H, Hudry-Clergeon, G, Petillot, Y, Norager, S, Medina, M, Frey, M. | | Deposit date: | 1999-07-01 | | Release date: | 1999-12-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Refined X-ray structures of the oxidized, at 1.3 A, and reduced, at 1.17 A, [2Fe-2S] ferredoxin from the cyanobacterium Anabaena PCC7119 show redox-linked conformational changes.
Biochemistry, 38, 1999
|
|
