4XMK
 
 | |
2DBQ
 
 | | Crystal Structure of Glyoxylate Reductase (PH0597) from Pyrococcus horikoshii OT3, Complexed with NADP (I41) | | Descriptor: | GLYCEROL, Glyoxylate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Yoshikawa, S, Arai, R, Kinoshita, Y, Uchikubo-Kamo, T, Akasaka, R, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-16 | | Release date: | 2006-06-16 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of archaeal glyoxylate reductase from Pyrococcus horikoshii OT3 complexed with nicotinamide adenine dinucleotide phosphate.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2DBZ
 
 | | Crystal Structure of Glyoxylate Reductase (PH0597) from Pyrococcus horikoshii OT3, Complexed with NADP (P61) | | Descriptor: | Glyoxylate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Yoshikawa, S, Arai, R, Kinoshita, Y, Uchikubo-Kamo, T, Akasaka, R, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-16 | | Release date: | 2006-06-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of archaeal glyoxylate reductase from Pyrococcus horikoshii OT3 complexed with nicotinamide adenine dinucleotide phosphate.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1IMA
 
 | |
8TOS
 
 | | ACE2-peptide 6 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Christie, M, Payne, R. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
2DBR
 
 | | Crystal Structure of Glyoxylate Reductase (PH0597) from Pyrococcus horikoshii OT3, Complexed with NADP (P1) | | Descriptor: | Glyoxylate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Yoshikawa, S, Arai, R, Kinoshita, Y, Uchikubo-Kamo, T, Akasaka, R, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-16 | | Release date: | 2006-06-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of archaeal glyoxylate reductase from Pyrococcus horikoshii OT3 complexed with nicotinamide adenine dinucleotide phosphate.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
8C83
 
 | | Cryo-EM structure of in vitro reconstituted Otu2-bound Ub-40S complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S11-A, ... | | Authors: | Ikeuchi, K, Buschauer, R, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2023-01-18 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2.
Nat Commun, 14, 2023
|
|
8CAH
 
 | | Cryo-EM structure of native Otu2-bound ubiquitinated 43S pre-initiation complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Ikeuchi, K, Buschauer, R, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2023-01-24 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2.
Nat Commun, 14, 2023
|
|
1F5Z
 
 | |
1FEM
 
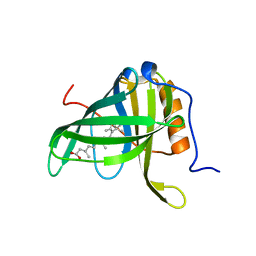 | | CRYSTALLOGRAPHIC STUDIES ON COMPLEXES BETWEEN RETINOIDS AND PLASMA RETINOL-BINDING PROTEIN | | Descriptor: | RETINOIC ACID, RETINOL BINDING PROTEIN | | Authors: | Zanotti, G, Marcello, M, Malpeli, G, Sartori, G, Berni, R. | | Deposit date: | 1994-08-29 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic studies on complexes between retinoids and plasma retinol-binding protein.
J.Biol.Chem., 269, 1994
|
|
1F6K
 
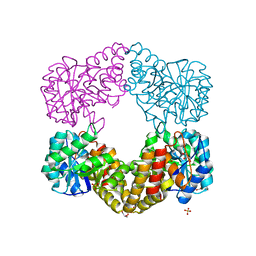 | | CRYSTAL STRUCTURE ANALYSIS OF N-ACETYLNEURAMINATE LYASE FROM HAEMOPHILUS INFLUENZAE: CRYSTAL FORM II | | Descriptor: | GLYCEROL, N-ACETYLNEURAMINATE LYASE, SULFATE ION | | Authors: | Barbosa, J.A.R.G, Smith, B.J, DeGori, R, Lawrence, M.C. | | Deposit date: | 2000-06-21 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Active site modulation in the N-acetylneuraminate lyase sub-family as revealed by the structure of the inhibitor-complexed Haemophilus influenzae enzyme.
J.Mol.Biol., 303, 2000
|
|
6X7H
 
 | |
5IIT
 
 | |
4XML
 
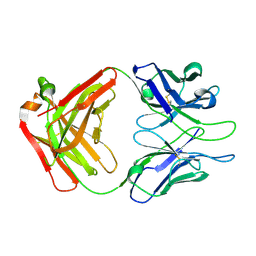 | | Crystal structure of Fab of HIV-1 gp120 V3-specific human monoclonal antibody 2424 | | Descriptor: | Heavy chain of HIV-1 gp120 V3-specific human monoclonal antibody 2424, Light chain of HIV-1 gp120 V3-specific human monoclonal antibody 2424 | | Authors: | Pan, R, Kong, X.-P. | | Deposit date: | 2015-01-14 | | Release date: | 2015-07-08 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Functional and Structural Characterization of Human V3-Specific Monoclonal Antibody 2424 with Neutralizing Activity against HIV-1 JRFL.
J.Virol., 89, 2015
|
|
1F6P
 
 | |
8PIX
 
 | | Cryo EM structure of the type 3C polymorph of alpha-synuclein at low pH. | | Descriptor: | Alpha-synuclein | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-22 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation
Elife, 2023
|
|
5IJH
 
 | |
1F74
 
 | | CRYSTAL STRUCTURE ANALYSIS OF N-ACETYLNEURAMINATE LYASE FROM HAEMOPHILUS INFLUENZAE: CRYSTAL FORM II COMPLEXED WITH 4-DEOXY-SIALIC ACID | | Descriptor: | 6,7,8,9-TETRAHYDROXY-5-METHYLCARBOXAMIDO-2-OXONONANOIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Barbosa, J.A.R.G, Smith, B.J, DeGori, R, Lawrence, M.C. | | Deposit date: | 2000-06-26 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Active site modulation in the N-acetylneuraminate lyase sub-family as revealed by the structure of the inhibitor-complexed Haemophilus influenzae enzyme.
J.Mol.Biol., 303, 2000
|
|
1F7B
 
 | | CRYSTAL STRUCTURE ANALYSIS OF N-ACETYLNEURAMINATE LYASE FROM HAEMOPHILUS INFLUENZAE: CRYSTAL FORM II IN COMPLEX WITH 4-OXO-SIALIC ACID | | Descriptor: | 4,4,6,7,8,9-HEXAHYDROXY-5-METHYLCARBOXAMIDONONANOIC ACID, 6,7,8,9-TETRAHYDROXY-5-METHYLCARBOXAMIDO-4-OXONONANOIC ACID, CHLORIDE ION, ... | | Authors: | Barbosa, J.A.R.G, Smith, B.J, DeGori, R, Lawrence, M.C. | | Deposit date: | 2000-06-26 | | Release date: | 2000-11-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Active site modulation in the N-acetylneuraminate lyase sub-family as revealed by the structure of the inhibitor-complexed Haemophilus influenzae enzyme.
J.Mol.Biol., 303, 2000
|
|
1F73
 
 | | CRYSTAL STRUCTURE ANALYSIS OF N-ACETYLNEURAMINATE LYASE FROM HAEMOPHILUS INFLUENZAE: CRYSTAL FORM III IN COMPLEX WITH SIALIC ACID ALDITOL | | Descriptor: | 2,4,6,7,8,9-HEXAHYDROXY-5-METHYLCARBOXAMIDO NONANOIC ACID, GLYCEROL, N-ACETYL NEURAMINATE LYASE | | Authors: | Barbosa, J.A.R.G, Smith, B.J, DeGori, R, Lawrence, M.C. | | Deposit date: | 2000-06-25 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Active site modulation in the N-acetylneuraminate lyase sub-family as revealed by the structure of the inhibitor-complexed Haemophilus influenzae enzyme.
J.Mol.Biol., 303, 2000
|
|
4Y1W
 
 | |
5BX5
 
 | | Crystal structure of Thermoanaerobacterium xylanolyticum GH116 beta-glucosidase with glucose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Charoenwattanasatien, R, Pengthaisong, S, Sansenya, S, Mutoh, R, Hua, Y, Tanaka, H, Kurisu, G, Ketudat Cairns, J.R. | | Deposit date: | 2015-06-08 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bacterial beta-Glucosidase Reveals the Structural and Functional Basis of Genetic Defects in Human Glucocerebrosidase 2 (GBA2)
Acs Chem.Biol., 11, 2016
|
|
5BX4
 
 | | Crystal structure of Thermoanaerobacterium xylanolyticum GH116 beta-glucosidase with Glucoimidazole | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLUCOIMIDAZOLE, ... | | Authors: | Charoenwattanasatien, R, Pengthaisong, S, Sansenya, S, Mutoh, R, Tankrathok, A, Tanaka, H, Kurisu, G, Ketudat Cairns, J.R. | | Deposit date: | 2015-06-08 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Bacterial beta-Glucosidase Reveals the Structural and Functional Basis of Genetic Defects in Human Glucocerebrosidase 2 (GBA2)
Acs Chem.Biol., 11, 2016
|
|
5IDK
 
 | |
5BX2
 
 | | Crystal structure of Thermoanaerobacterium xylanolyticum GH116 beta-glucosidase with 2-deoxy-2-fluoroglucoside | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-2-fluoro-alpha-D-glucopyranose, CALCIUM ION, ... | | Authors: | Charoenwattanasatien, R, Pengthaisong, S, Sansenya, S, Mutoh, R, Tanaka, H, Kurisu, G, Ketudat Cairns, J.R. | | Deposit date: | 2015-06-08 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Bacterial beta-Glucosidase Reveals the Structural and Functional Basis of Genetic Defects in Human Glucocerebrosidase 2 (GBA2)
Acs Chem.Biol., 11, 2016
|
|
