6PF0
 
 | |
6PEZ
 
 | |
5MSZ
 
 | | Lytic Polysaccharide Monooxygenase AA15 from Thermobia domestica in the Cu(I) State | | Descriptor: | 1,2-ETHANEDIOL, COPPER (I) ION, Thermobia domestica domestica AA15 | | Authors: | Hemsworth, G.R, Sabbadin, F, Ciano, L, Henrissat, B, Dupree, P, Tryfona, T, Besser, K, Elias, L, Pesante, G, Li, Y, Dowle, A, Bates, R, Gomez, L, Hallam, R, Davies, G.J, Walton, P.H, Bruce, N.C, McQueen-Mason, S. | | Deposit date: | 2017-01-06 | | Release date: | 2018-02-28 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | An ancient family of lytic polysaccharide monooxygenases with roles in arthropod development and biomass digestion.
Nat Commun, 9, 2018
|
|
6WLL
 
 | | Apo F. nucleatum glycine riboswitch models, 10.0 Angstrom resolution | | Descriptor: | RNA (171-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
7T9U
 
 | | Crystal structure of hSTING with an agonist (SHR169224) | | Descriptor: | (3S,4S)-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-4-(prop-2-en-1-yl)-4,5-dihydroimidazo[1,5,4-de][1,4]benzoxazine-8-carboxamide, CALCIUM ION, Stimulator of interferon genes protein | | Authors: | Chowdhury, R, Miller, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | SHR1032, a novel STING agonist, stimulates anti-tumor immunity and directly induces AML apoptosis.
Sci Rep, 12, 2022
|
|
6WLJ
 
 | | ATP-TTR-3 with AMP models, 9.6 Angstrom resolution | | Descriptor: | RNA (130-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
7T9V
 
 | | Crystal structure of hSTING with the agonist, SHR171032 | | Descriptor: | (3S,4S)-4-(3-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methoxy-1H-benzimidazol-1-yl}propyl)-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-4,5-dihydroimidazo[1,5,4-de][1,4]benzoxazine-8-carboxamide, CALCIUM ION, Stimulator of interferon genes protein | | Authors: | Chowdhury, R, Miller, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | SHR1032, a novel STING agonist, stimulates anti-tumor immunity and directly induces AML apoptosis.
Sci Rep, 12, 2022
|
|
6YCC
 
 | | Structure the ananain protease from Ananas comosus covalently bound to the E64 inhibitor | | Descriptor: | Ananain, GLYCEROL, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
6PK8
 
 | | Antibody scFv-M204 dimeric state | | Descriptor: | SULFATE ION, scFv-M204 antibody | | Authors: | Abskharon, R, Sawaya, M.R, Seidler, P.M, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2019-06-28 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structure of a conformational antibody that binds tau oligomers and inhibits pathological seeding by extracts from donors with Alzheimer's disease.
J.Biol.Chem., 295, 2020
|
|
6YL3
 
 | | High resolution cryo-EM structure of urease from the pathogen Yersinia enterocolitica | | Descriptor: | NICKEL (II) ION, Urease subunit alpha, Urease subunit beta, ... | | Authors: | Righetto, R.D, Anton, L, Adaixo, R, Jakob, R, Zivanov, J, Mahi, M.A, Ringler, P, Schwede, T, Maier, T, Stahlberg, H. | | Deposit date: | 2020-04-06 | | Release date: | 2020-05-06 | | Last modified: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (1.98 Å) | | Cite: | High-resolution cryo-EM structure of urease from the pathogen Yersinia enterocolitica.
Nat Commun, 11, 2020
|
|
1BVN
 
 | | PIG PANCREATIC ALPHA-AMYLASE IN COMPLEX WITH THE PROTEINACEOUS INHIBITOR TENDAMISTAT | | Descriptor: | CALCIUM ION, CHLORIDE ION, PROTEIN (ALPHA-AMYLASE), ... | | Authors: | Machius, M, Wiegand, G, Epp, O, Huber, R. | | Deposit date: | 1998-09-16 | | Release date: | 1998-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of porcine pancreatic alpha-amylase in complex with the microbial inhibitor Tendamistat.
J.Mol.Biol., 247, 1995
|
|
6WLK
 
 | | Apo ATP-TTR-3 models, 10.0 Angstrom resolution | | Descriptor: | RNA (130-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLU
 
 | | V. cholerae glycine riboswitch with glycine models, 5.7 Angstrom resolution | | Descriptor: | RNA (231-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
1BZW
 
 | | PEANUT LECTIN COMPLEXED WITH C-LACTOSE | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PROTEIN (PEANUT LECTIN), ... | | Authors: | Ravishankar, R, Surolia, A, Vijayan, M, Lim, S, Kishi, Y. | | Deposit date: | 1998-11-05 | | Release date: | 1998-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Preferred Conformation of C-Lactose at the Free and Peanut Lectin Bound States
J.Am.Chem.Soc., 120, 1998
|
|
8API
 
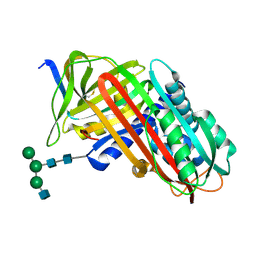 | | THE S VARIANT OF HUMAN ALPHA1-ANTITRYPSIN, STRUCTURE AND IMPLICATIONS FOR FUNCTION AND METABOLISM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-1 ANTITRYPSIN (CHAIN A), ... | | Authors: | Loebermann, H, Tokuoka, R, Deisenhofer, J, Huber, R. | | Deposit date: | 1988-09-08 | | Release date: | 1990-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The S variant of human alpha 1-antitrypsin, structure and implications for function and metabolism.
Protein Eng., 2, 1989
|
|
7GN7
 
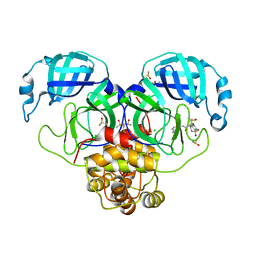 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-1cbc2fae-2 (Mpro-P2381) | | Descriptor: | (4S)-6-chloro-4-ethyl-N-(isoquinolin-4-yl)-1,1-dioxo-1,2,3,4-tetrahydro-1lambda~6~,2-benzothiazine-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GL7
 
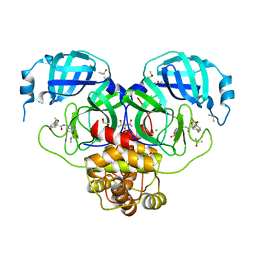 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-6bf93aa8-1 (Mpro-P1783) | | Descriptor: | (4S)-6-chloro-4-methoxy-N-[7-(methylsulfamoyl)isoquinolin-4-yl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
6XE9
 
 | | 10S myosin II (smooth muscle) | | Descriptor: | Myosin II heavy chain (smooth muscle), Myosin light chain 9, Myosin light chain smooth muscle isoform | | Authors: | Tiwari, P, Craig, R, Padron, R. | | Deposit date: | 2020-06-12 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of the inhibited (10S) form of myosin II.
Nature, 588, 2020
|
|
7GNM
 
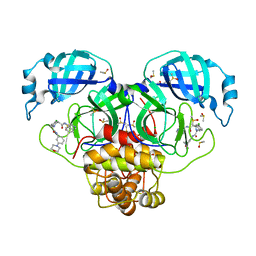 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with NIR-WEI-dcc3321b-2 (Mpro-P2761) | | Descriptor: | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-{2-[3-(morpholin-4-yl)anilino]-2-oxoethyl}-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GIK
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-1f3f1a6f-1 (Mpro-P0098) | | Descriptor: | (2R)-2-amino-2-(3,4-dichlorophenyl)-N-(isoquinolin-4-yl)acetamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GLO
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with JAN-GHE-5a013bed-2 (Mpro-P2001) | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.927 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GJ0
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDG-MED-70ae9412-2 (Mpro-P0151) | | Descriptor: | (4R)-6-chloro-4-{[(N,N-dimethylglycyl)amino]methyl}-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GM4
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-6f6ae286-5 (Mpro-P2080) | | Descriptor: | (4S)-6-chloro-N-(7-fluoroisoquinolin-4-yl)-2-[2-(methylamino)-2-oxoethyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GJG
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-UNI-8e43a71e-8 (Mpro-P0238) | | Descriptor: | (4S)-6-chloro-4-{2-[4-(3-hydroxypropyl)piperazin-1-yl]-2-oxoethyl}-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GMJ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-477dc5b7-4 (Mpro-P2197) | | Descriptor: | (3'R)-6-chloro-1'-(isoquinolin-4-yl)-2,3-dihydrospiro[[1]benzopyran-4,3'-pyrrolidin]-2'-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
