1F0Q
 
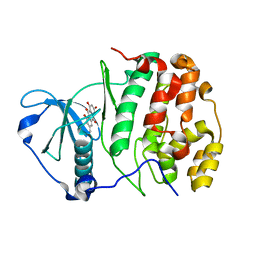 | | CRYSTAL STRUCTURE OF THE ALPHA SUBUNIT OF PROTEIN KINASE CK2 IN COMPLEX WITH THE NUCLEOTIDE COMPETITIVE INHIBITOR EMODIN | | Descriptor: | 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, PROTEIN KINASE CK2, ALPHA SUBUNIT | | Authors: | Battistutta, R, Sarno, S, De Moliner, E, Papinutto, E, Zanotti, G, Pinna, L.A. | | Deposit date: | 2000-05-17 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | The replacement of ATP by the competitive inhibitor emodin induces conformational modifications in the catalytic site of protein kinase CK2.
J.Biol.Chem., 275, 2000
|
|
6OXT
 
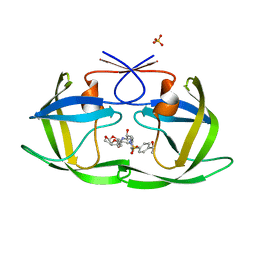 | | HIV-1 Protease NL4-3 WT in Complex with LR-84 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{({4-[(1S)-1-hydroxyethyl]phenyl}sulfonyl)[(2S)-2-methylbutyl]amino}-1-phenylbutan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OY0
 
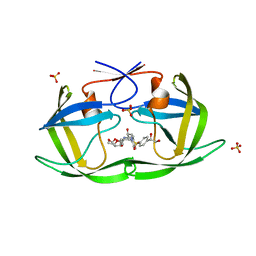 | | HIV-1 Protease NL4-3 WT in Complex with LR2-21 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{({4-[(1S)-1,2-dihydroxyethyl]phenyl}sulfonyl)[(2S)-2-methylbutyl]amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
4XJM
 
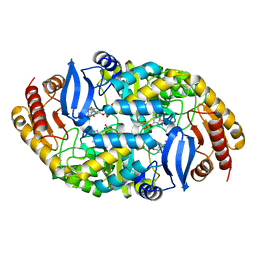 | |
6OMX
 
 | |
7T8I
 
 | |
7PL5
 
 | | Crystal structure of choline-binding module (R1-R9) of LytB from Streptococcus pneumoniae | | Descriptor: | CHOLINE ION, Putative endo-beta-N-acetylglucosaminidase, TRIETHYLENE GLYCOL, ... | | Authors: | Molina, R, Martinez Caballero, S, Hermoso, J.A. | | Deposit date: | 2021-08-28 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular basis of the final step of cell division in Streptococcus pneumoniae.
Cell Rep, 42, 2023
|
|
7T4Q
 
 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with neutralizing fabs 2C12, 7I13 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
8SGZ
 
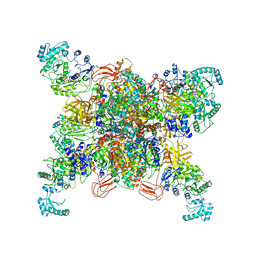 | | Leishmania tarentolae propionyl-CoA carboxylase (alpha-6-beta-6) | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Propionyl-coa carboxylase beta chain, putative, ... | | Authors: | Lee, J.K.J, Liu, Y.T, Hu, J.J, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2023-04-13 | | Release date: | 2023-05-17 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | CryoEM reveals oligomeric isomers of a multienzyme complex and assembly mechanics.
J Struct Biol X, 7, 2023
|
|
7T4R
 
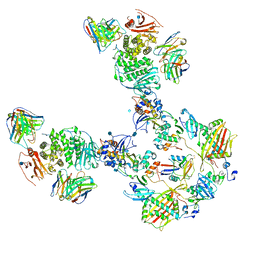 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with THBD and neutralizing fabs MSL-109 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
8SGX
 
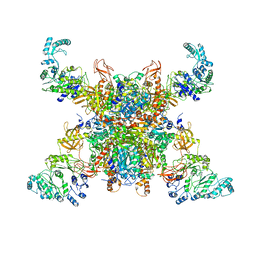 | | Leishmania tarentolae propionyl-CoA carboxylase (alpha-4-beta-6) | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Propionyl-coa carboxylase beta chain, putative, ... | | Authors: | Lee, J.K.J, Liu, Y.T, Hu, J.J, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2023-04-13 | | Release date: | 2023-05-17 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (10.3 Å) | | Cite: | CryoEM reveals oligomeric isomers of a multienzyme complex and assembly mechanics.
J Struct Biol X, 7, 2023
|
|
8SGY
 
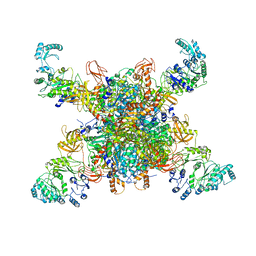 | | Leishmania tarentolae propionyl-CoA carboxylase (alpha-5-beta-6) | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Propionyl-coa carboxylase beta chain, putative, ... | | Authors: | Lee, J.K.J, Liu, Y.T, Hu, J.J, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2023-04-13 | | Release date: | 2023-05-17 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (8.62 Å) | | Cite: | CryoEM reveals oligomeric isomers of a multienzyme complex and assembly mechanics.
J Struct Biol X, 7, 2023
|
|
7T4S
 
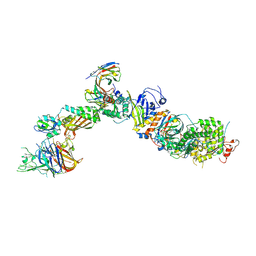 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with NRP2 and neutralizing fabs 8I21 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
1F50
 
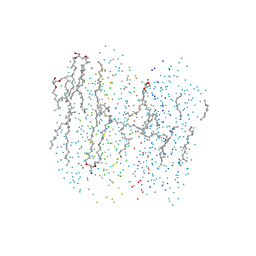 | | BACTERIORHODOPSIN-BR STATE OF THE E204Q MUTANT AT 1.7 ANGSTROM RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, BACTERIORHODOPSIN, ... | | Authors: | Luecke, H, Schobert, B, Cartailler, J.P, Richter, H.T, Rosengarth, A, Needleman, R, Lanyi, J.K. | | Deposit date: | 2000-06-10 | | Release date: | 2000-08-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Coupling photoisomerization of retinal to directional transport in bacteriorhodopsin.
J.Mol.Biol., 300, 2000
|
|
4XMU
 
 | |
8SIK
 
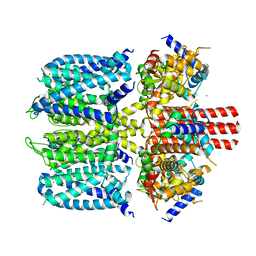 | | KCNQ1 with voltage sensor in the up conformation | | Descriptor: | CALCIUM ION, Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Mandala, V.S, MacKinnon, R. | | Deposit date: | 2023-04-16 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The membrane electric field regulates the PIP 2 -binding site to gate the KCNQ1 channel.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4XMZ
 
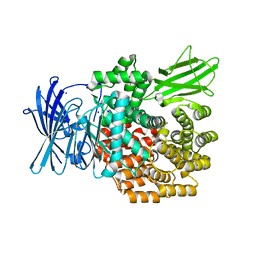 | |
1F56
 
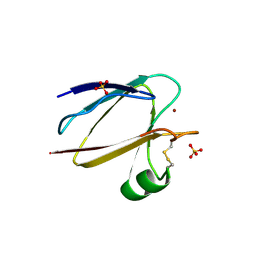 | | SPINACH PLANTACYANIN | | Descriptor: | COPPER (I) ION, PLANTACYANIN, SULFATE ION | | Authors: | Einsle, O, Mehrabian, Z, Nalbandyan, R, Messerschmidt, A. | | Deposit date: | 2000-06-13 | | Release date: | 2000-11-01 | | Last modified: | 2021-07-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of plantacyanin, a basic blue cupredoxin from spinach.
J.Biol.Inorg.Chem., 5, 2000
|
|
7SP1
 
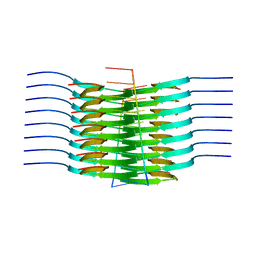 | | RNA-induced tau amyloid fibril | | Descriptor: | Isoform Tau-F of Microtubule-associated protein tau, RNA (5'-R(*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3') | | Authors: | Abskharon, R, Sawaya, M.R, Boyer, D.R, Eisenberg, D.S. | | Deposit date: | 2021-11-02 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of RNA-induced tau fibrils reveals a small C-terminal core that may nucleate fibril formation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4XN9
 
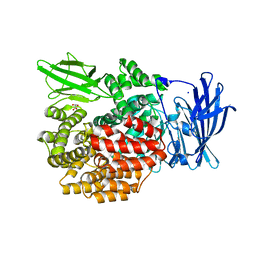 | |
7SZ6
 
 | | Kinetically trapped Pseudomonas-phage PaP3 portal protein - delta barrel mutant class-3 | | Descriptor: | Portal protein | | Authors: | Hou, C.-F.D, Swanson, N.A, Li, F, Yang, R, Lokareddy, R.K, Cingolani, G. | | Deposit date: | 2021-11-25 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (6.24 Å) | | Cite: | Cryo-EM Structure of a Kinetically Trapped Dodecameric Portal Protein from the Pseudomonas-phage PaP3.
J.Mol.Biol., 434, 2022
|
|
4XOC
 
 | | Crystal structure of the FimH lectin domain from E.coli F18 in complex with heptyl alpha-D-mannopyrannoside | | Descriptor: | FimH protein, heptyl alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Sauer, M.M, Navarra, G, Ernst, B, Glockshuber, R, Maier, T. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Catch-bond mechanism of the bacterial adhesin FimH.
Nat Commun, 7, 2016
|
|
8SIM
 
 | | KCNQ1 with voltage sensor in the intermediate conformation | | Descriptor: | CALCIUM ION, Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Mandala, V.S, MacKinnon, R. | | Deposit date: | 2023-04-16 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | The membrane electric field regulates the PIP 2 -binding site to gate the KCNQ1 channel.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8SIN
 
 | | KCNQ1 with voltage sensor in the down conformation | | Descriptor: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Mandala, V.S, MacKinnon, R. | | Deposit date: | 2023-04-16 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | The membrane electric field regulates the PIP 2 -binding site to gate the KCNQ1 channel.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7SZ4
 
 | | Kinetically trapped Pseudomonas-phage PaP3 portal protein - delta barrel mutant class-2 | | Descriptor: | Portal protein | | Authors: | Hou, C.-F.D, Swanson, N.A, Li, F, Yang, R, Lokareddy, R.K, Cingolani, G. | | Deposit date: | 2021-11-25 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM Structure of a Kinetically Trapped Dodecameric Portal Protein from the Pseudomonas-phage PaP3.
J.Mol.Biol., 434, 2022
|
|
