1B9G
 
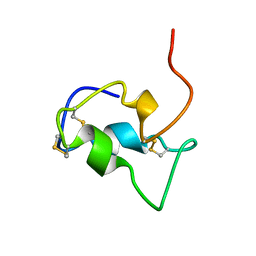 | | INSULIN-LIKE-GROWTH-FACTOR-1 | | Descriptor: | PROTEIN (GROWTH FACTOR IGF-1) | | Authors: | De Wolf, E, Gill, R, Geddes, S, Pitts, J, Wollmer, A, Grotzinger, J. | | Deposit date: | 1999-02-11 | | Release date: | 1999-02-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a mini IGF-1.
Protein Sci., 5, 1996
|
|
1J2E
 
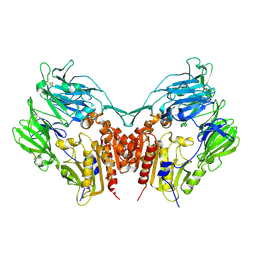 | | Crystal structure of Human Dipeptidyl peptidase IV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase IV | | Authors: | Hiramatsu, H, Kyono, K, Higashiyama, Y, Fukushima, C, Shima, H, Sugiyama, S, Inaka, K, Yamamoto, A, Shimizu, R. | | Deposit date: | 2002-12-30 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure and function of human dipeptidyl peptidase IV, possessing a unique eight-bladed beta-propeller fold.
Biochem.Biophys.Res.Commun., 302, 2003
|
|
2WTV
 
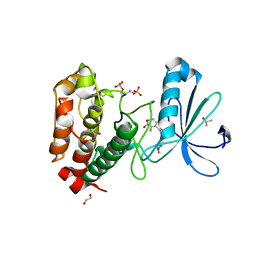 | | Aurora-A Inhibitor Structure | | Descriptor: | 1,2-ETHANEDIOL, 4-{[9-CHLORO-7-(2,6-DIFLUOROPHENYL)-5H-PYRIMIDO[5,4-D][2]BENZAZEPIN-2-YL]AMINO}BENZOIC ACID, ACETATE ION, ... | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2009-09-22 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of an Aurora-A Mutant that Mimics Aurora-B Bound to Mln8054: Insights Into Selectivity and Drug Design.
Biochem.J., 427, 2010
|
|
4BG4
 
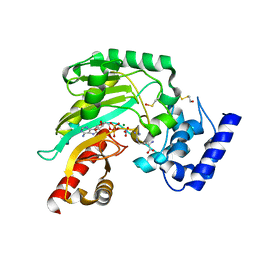 | | Crystal structure of Litopenaeus vannamei arginine kinase in a ternary analog complex with arginine, ADP-Mg and NO3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, ARGININE KINASE, ... | | Authors: | Lopez-Zavala, A.A, Garcia-Orozco, K.D, Carrasco-Miranda, J.S, Sugich-Miranda, R, Velazquez-Contreras, E.F, Criscitiello, M.F, GBrieba, L, Rudino-Pinera, E, Sotelo-Mundo, R.R. | | Deposit date: | 2013-03-22 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystal Structure of Shrimp Arginine Kinase in Binary Complex with Arginine-A Molecular View of the Phosphagen Precursor Binding to the Enzyme.
J.Bioenerg.Biomembr., 45, 2013
|
|
2X9X
 
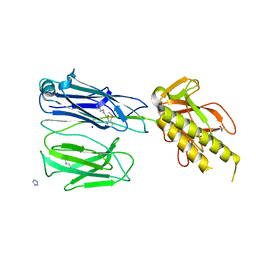 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN, IMIDAZOLE, SODIUM ION | | Authors: | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
1BC7
 
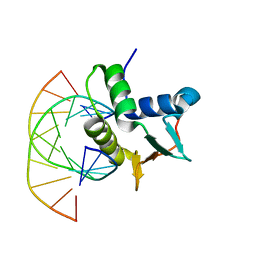 | | SERUM RESPONSE FACTOR ACCESSORY PROTEIN 1A (SAP-1)/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*AP*CP*AP*TP*CP*CP*TP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*AP*GP*GP*AP*TP*GP*TP*G)-3'), PROTEIN (ETS-DOMAIN PROTEIN) | | Authors: | Mo, Y, Vaessen, B, Johnston, K, Marmorstein, R. | | Deposit date: | 1998-05-05 | | Release date: | 1999-01-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structures of SAP-1 bound to DNA targets from the E74 and c-fos promoters: insights into DNA sequence discrimination by Ets proteins.
Mol.Cell, 2, 1998
|
|
2WZY
 
 | | Crystal structure of A-AChBP in complex with 13-desmethyl spirolide C | | Descriptor: | 13-DESMETHYL SPIROLIDE C, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Bourne, Y, Radic, Z, Araoz, R, Talley, T.T, Benoit, E, Servent, D, Taylor, P, Molgo, J, Marchot, P. | | Deposit date: | 2009-12-03 | | Release date: | 2010-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural Determinants in Phycotoxins and Achbp Conferring High Affinity Binding and Nicotinic Achr Antagonism.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2WUT
 
 | | Crystal structure of human myelin protein P2 in complex with palmitate | | Descriptor: | CHLORIDE ION, GLYCEROL, MYELIN P2 PROTEIN, ... | | Authors: | Majava, V, Nanekar, R, Kursula, P. | | Deposit date: | 2009-10-09 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Functional Characterization of Human Peripheral Nervous System Myelin Protein P2.
Plos One, 5, 2010
|
|
1J2J
 
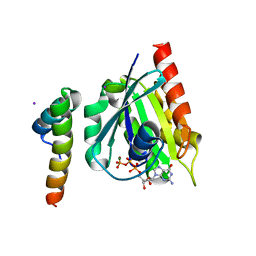 | | Crystal structure of GGA1 GAT N-terminal region in complex with ARF1 GTP form | | Descriptor: | ADP-ribosylation factor 1, ADP-ribosylation factor binding protein GGA1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shiba, T, Kawasaki, M, Takatsu, H, Nogi, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2003-01-05 | | Release date: | 2003-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport
NAT.STRUCT.BIOL., 10, 2003
|
|
4BF1
 
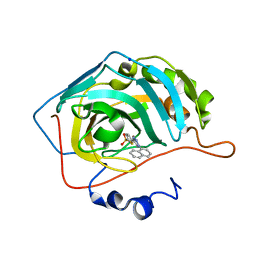 | | Three dimensional structure of human carbonic anhydrase II in complex with 5-(1-naphthalen-1-yl-1,2,3-triazol-4-yl)thiophene-2-sulfonamide | | Descriptor: | 5-(1-naphthalen-1-yl-1,2,3-triazol-4-yl)thiophene-2-sulfonamide, CARBONIC ANHYDRASE 2, SODIUM ION, ... | | Authors: | Tars, K, Leitans, J, Zalubovskis, R. | | Deposit date: | 2013-03-13 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | 5-Substituted-(1,2,3-Triazol-4-Yl)Thiophene-2-Sulfonamides Strongly Inhibit Human Carbonic Anhydrases I, II, Ix and Xii: Solution and X-Ray Crystallographic Studies.
Bioorg.Med.Chem., 21, 2013
|
|
4BK2
 
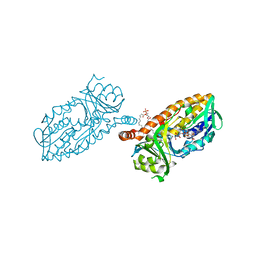 | | Crystal structure of 3-hydroxybenzoate 6-hydroxylase uncovers lipid- assisted flavoprotein strategy for regioselective aromatic hydroxylation: Q301E mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATIDYLGLYCEROL-PHOSPHOGLYCEROL, PROBABLE SALICYLATE MONOOXYGENASE | | Authors: | Orru, R, Montersino, S, Barendregt, A, Westphal, A.H, van Duijn, E, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2013-04-21 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal Structure of 3-Hydroxybenzoate 6-Hydroxylase Uncovers Lipid-Assisted Flavoprotein Strategy for Regioselective Aromatic Hydroxylation
J.Biol.Chem., 288, 2013
|
|
2X1S
 
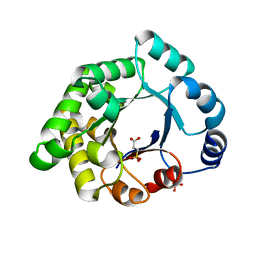 | | Crystallographic binding studies with an engineered monomeric variant of triosephosphate isomerase | | Descriptor: | 3-SULFOPROPANOIC ACID, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE, ... | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Castejeijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-04 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1J95
 
 | |
4ARP
 
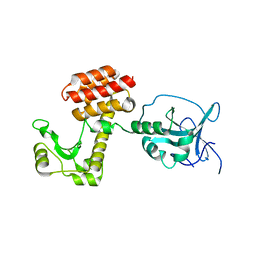 | | Structure of the inactive pesticin E178A mutant | | Descriptor: | PESTICIN | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
2WZT
 
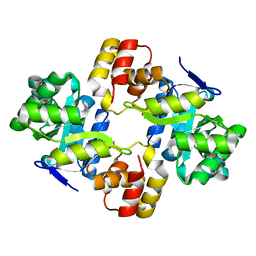 | | Crystal structure of a mycobacterium aldo-keto reductase in its apo and liganded form | | Descriptor: | ALDO-KETO REDUCTASE | | Authors: | Scoble, J, McAlister, A.D, Fulton, Z, Troy, S, Byres, E, Vivian, J.P, Brammananth, R, Wilce, M.C.J, Le Nours, J, Zaker-Tabrizi, L, Coppel, R.L, Crellin, P.K, Rossjohn, J, Beddoe, T. | | Deposit date: | 2009-12-03 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure and Comparative Functional Analyses of a Mycobacterium Aldo-Keto Reductase.
J.Mol.Biol., 398, 2010
|
|
2X09
 
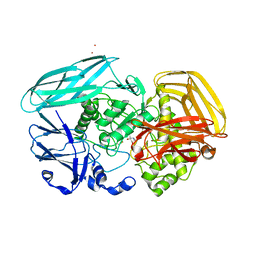 | | Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue | | Descriptor: | AMINO-AUSTRALINE, CADMIUM ION, EXO-BETA-D-GLUCOSAMINIDASE | | Authors: | Pluvinage, B, Ghinet, M.G, Brzezinski, R, Boraston, A.B, Stubbs, K.A. | | Deposit date: | 2009-12-07 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of the Exo-Beta-D-Glucosaminidase Csxa by a Glucosamine-Configured Castanospermine and an Amino-Australine Analogue.
Org.Biomol.Chem., 7, 2009
|
|
4B3U
 
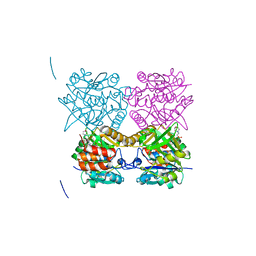 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-amino-1-benzyl-5-(ethylamino)pyrimidine-2,4(1H,3H)-dione, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Sarkar, A, Brenk, R, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-07-26 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
1B90
 
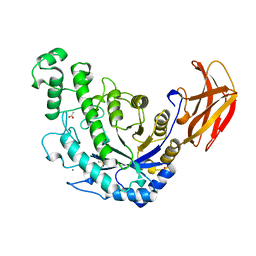 | | BACILLUS CEREUS BETA-AMYLASE APO FORM | | Descriptor: | ACETATE ION, CALCIUM ION, PROTEIN (BETA-AMYLASE), ... | | Authors: | Mikami, B, Adachi, M, Kage, T, Sarikaya, E, Nanmori, T, Shinke, R, Utsumi, S. | | Deposit date: | 1999-03-06 | | Release date: | 1999-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of raw starch-digesting Bacillus cereus beta-amylase complexed with maltose.
Biochemistry, 38, 1999
|
|
5QHL
 
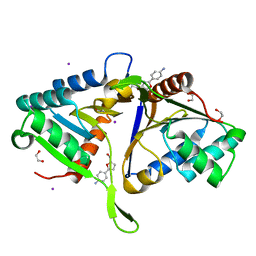 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human FAM83B in complex with FMOPL000551a | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, Protein FAM83B, ... | | Authors: | Pinkas, D.M, Bufton, J.C, Fox, A.E, Talon, R, Krojer, T, Douangamath, A, Collins, P, Zhang, R, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Bullock, A.N. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QI9
 
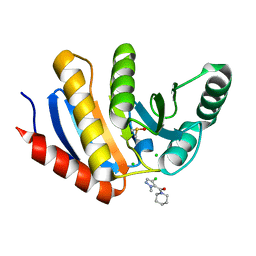 | | PanDDA analysis group deposition -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000711a | | Descriptor: | (4-chloranyl-2-methyl-pyrazol-3-yl)-piperidin-1-yl-methanone, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QIO
 
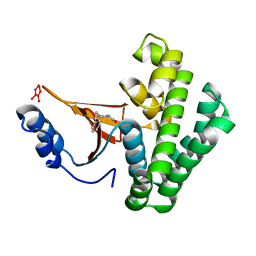 | | Covalent fragment group deposition -- Crystal Structure of OUTB2 in complex with P11 | | Descriptor: | (1S,2S)-N'-(chloroacetyl)-2-phenylcyclopropane-1-carbohydrazide, UNKNOWN LIGAND, Ubiquitin thioesterase OTUB2 | | Authors: | Sethi, R, Douangamath, A, Resnick, E, Bradley, A.R, Collins, P, Brandao-Neto, J, Talon, R, Krojer, T, Bountra, C, Arrowsmith, C.H, Edwards, A, London, N, von Delft, F. | | Deposit date: | 2018-08-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Covalent fragment group deposition
To Be Published
|
|
5QIS
 
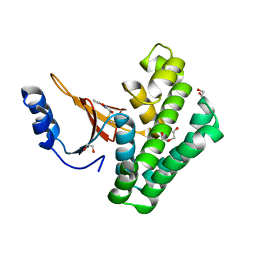 | | Covalent fragment group deposition -- Crystal Structure of OUTB2 in complex with PCM-0102500 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, N-(5-methyl-1,2-oxazol-3-yl)acetamide, ... | | Authors: | Sethi, R, Douangamath, A, Resnick, E, Bradley, A.R, Collins, P, Brandao-Neto, J, Talon, R, Krojer, T, Bountra, C, Arrowsmith, C.H, Edwards, A, London, N, von Delft, F. | | Deposit date: | 2018-08-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Covalent fragment group deposition
To Be Published
|
|
4A97
 
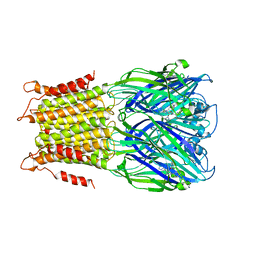 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with zopiclone | | Descriptor: | (5R)-6-(5-chloropyridin-2-yl)-7-oxo-6,7-dihydro-5H-pyrrolo[3,4-b]pyrazin-5-yl 4-methylpiperazine-1-carboxylate, CYS-LOOP LIGAND-GATED ION CHANNEL | | Authors: | Spurny, R, Brams, M, Ulens, C. | | Deposit date: | 2011-11-24 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.343 Å) | | Cite: | Pentameric Ligand-Gated Ion Channel Elic is Activated by Gaba and Modulated by Benzodiazepines.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AOH
 
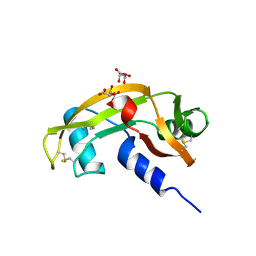 | | Structural snapshots and functional analysis of human angiogenin variants associated with Amyotrophic Lateral Sclerosis (ALS) | | Descriptor: | ANGIOGENIN, D(-)-TARTARIC ACID, L(+)-TARTARIC ACID | | Authors: | Thiyagarajan, N, Ferguson, R, Subramanian, V, Acharya, K.R. | | Deposit date: | 2012-03-27 | | Release date: | 2012-10-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.041 Å) | | Cite: | Structural and Molecular Insights Into the Mechanism of Action of Human Angiogenin-Als Variants in Neurons
Nat.Commun., 3, 2012
|
|
1J2B
 
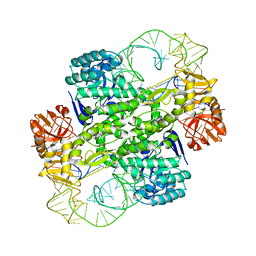 | | Crystal Structure Of Archaeosine tRNA-Guanine Transglycosylase Complexed With lambda-form tRNA(Val) | | Descriptor: | Archaeosine tRNA-guanine transglycosylase, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ishitani, R, Nureki, O, Nameki, N, Okada, N, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-29 | | Release date: | 2003-05-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Alternative Tertiary Structure of tRNA for Recognition by a Posttranscriptional Modification Enzyme
Cell(Cambridge,Mass.), 113, 2003
|
|
