3ZWN
 
 | | Crystal structure of Aplysia cyclase complexed with substrate NGD and product cGDPR | | Descriptor: | 3-(AMINOCARBONYL)-1-[(2R,3R,4S,5R)-5-({[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(2-AMINO-6-OXO-1,6-DIHYDRO-9H-PURIN-9-YL)-3,4-DIHYD ROXYTETRAHYDROFURAN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]OXY}METHYL)-3,4-DIHYDROXYTETRAHYDROFURAN-2- YL]PYRIDINIUM, ADP-RIBOSYL CYCLASE, CYCLIC GUANOSINE DIPHOSPHATE-RIBOSE | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-02 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
J.Mol.Biol., 415, 2012
|
|
3ZVE
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 84 | | Descriptor: | 3C PROTEASE, O-tert-butyl-N-[(9H-fluoren-9-ylmethoxy)carbonyl]-L-threonyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZVT
 
 | | Unexpected tricovalent binding mode of boronic acids within the active site of a penicillin binding protein | | Descriptor: | 2,6 DIMETHOXYBENZAMIDOBORONIC ACID, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, MAGNESIUM ION, ... | | Authors: | Sauvage, E, Zervosen, A, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | Deposit date: | 2011-07-27 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Unexpected Tricovalent Binding Mode of Boronic Acids within the Active Site of a Penicillin- Binding Protein.
J.Am.Chem.Soc., 133, 2011
|
|
6J4D
 
 | | Crystal structure of MarH, an epimerase for biosynthesis of Maremycins in Streptomyces, under pH 4.7, without Zn | | Descriptor: | CITRATE ANION, Cupin superfamily protein, GLYCEROL | | Authors: | Hou, Y, Liu, B, Hu, K, Zhang, R. | | Deposit date: | 2019-01-08 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis for the Isomerization Mechanism of MarH
To Be Published
|
|
3ZWX
 
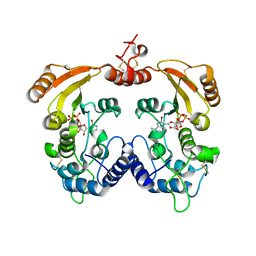 | | Crystal structure of ADP-ribosyl cyclase complexed with 8-bromo-ADP- ribose | | Descriptor: | ADP-RIBOSYL CYCLASE, CHLORIDE ION, [(2R,3S,4R,5R)-5-(6-amino-8-bromo-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3S,4S)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-03 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
To be Published
|
|
3ZCZ
 
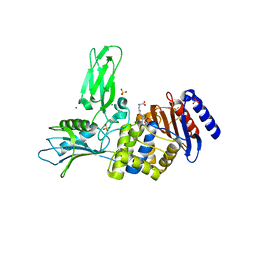 | | Crystal structure of a complex between Actinomadura R39 DD-peptidase and a trifluoroketone inhibitor | | Descriptor: | (2R)-2-amino-7-oxo-7-{[(2R,3S)-4,4,4-trifluoro-3-hydroxybutan-2-yl]amino}heptanoic acid, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, MAGNESIUM ION, ... | | Authors: | Sauvage, E, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | Deposit date: | 2012-11-23 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Inhibition of Dd-Peptidases by a Specific Trifluoroketone: Crystal Structure of a Complex with the Actinomadura R39 Dd-Peptidase.
Biochemistry, 52, 2013
|
|
7JLV
 
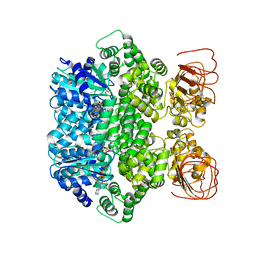 | | Structure of the activated Roq1 resistosome directly recognizing the pathogen effector XopQ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Disease resistance protein Roq1, MAGNESIUM ION | | Authors: | Martin, R, Qi, T, Zhang, H, Lui, F, King, M, Toth, C, Nogales, E, Staskawicz, B.J. | | Deposit date: | 2020-07-30 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ.
Science, 370, 2020
|
|
1RTJ
 
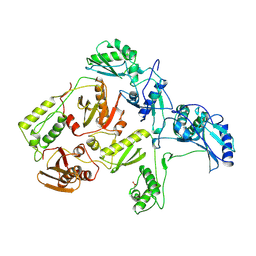 | | MECHANISM OF INHIBITION OF HIV-1 REVERSE TRANSCRIPTASE BY NON-NUCLEOSIDE INHIBITORS | | Descriptor: | HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Ross, C, Jones, Y, Stammers, D, Stuart, D. | | Deposit date: | 1995-05-03 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of inhibition of HIV-1 reverse transcriptase by non-nucleoside inhibitors.
Nat.Struct.Biol., 2, 1995
|
|
7JLT
 
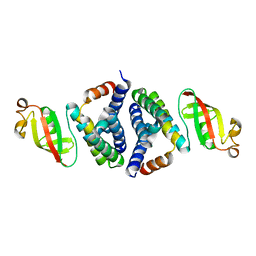 | | Crystal Structure of SARS-CoV-2 NSP7-NSP8 complex. | | Descriptor: | Non-structural protein 7, Non-structural protein 8 | | Authors: | Biswal, M, Hai, R, Song, J. | | Deposit date: | 2020-07-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two conserved oligomer interfaces of NSP7 and NSP8 underpin the dynamic assembly of SARS-CoV-2 RdRP.
Nucleic Acids Res., 49, 2021
|
|
3ZMZ
 
 | | LSD1-CoREST in complex with PRSFAV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-13 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
3RBG
 
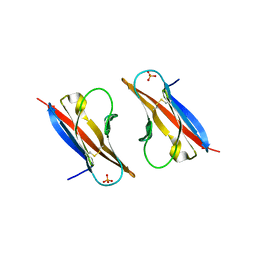 | | Crystal structure analysis of Class-I MHC restricted T-cell associated molecule | | Descriptor: | Cytotoxic and regulatory T-cell molecule, PHOSPHATE ION | | Authors: | Rubinstein, R, Ramagopal, U.A, Toro, R, Nathenson, S.G, Fiser, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2011-03-29 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional classification of immune regulatory proteins.
Structure, 21, 2013
|
|
3ZFR
 
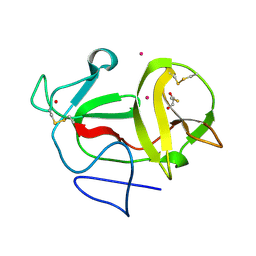 | | Crystal structure of product-like, processed N-terminal protease Npro with iridium | | Descriptor: | HYDROXIDE ION, IRIDIUM (III) ION, MONOTHIOGLYCEROL, ... | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZON
 
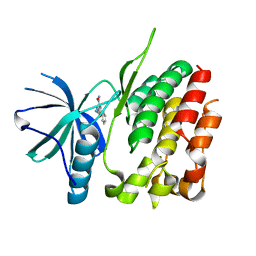 | | Human TYK2 pseudokinase domain bound to a kinase inhibitor | | Descriptor: | 5-PHENYL-2-UREIDOTHIOPHENE-3-CARBOXAMIDE, NON-RECEPTOR TYROSINE-PROTEIN KINASE TYK2 | | Authors: | Elkins, J.M, Wang, J, Krojer, T, Savitsky, P, Chalk, R, Daga, N, Salah, E, Berridge, G, Picaud, S, von Delft, F, Bountra, C, Edwards, A, Knapp, S. | | Deposit date: | 2013-02-22 | | Release date: | 2013-04-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Human Tyk2 Pseudokinase Domain Bound to a Kinase Inhibitor
To be Published
|
|
3ZOU
 
 | | Native structure of Farnesyl Pyrophosphate Synthase from Pseudomonas aeruginosa PA01, with bound fragment SPB02696, and substrate geranyl pyrophosphate. | | Descriptor: | 3-(2-oxo-1,3-benzoxazol-3(2H)-yl)propanoic acid, DIMETHYL SULFOXIDE, FARNESYL PYROPHOSPHATE SYNTHASE, ... | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2013-02-25 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Characterization of Substrate and Inhibitor Binding to Farnesyl Pyrophosphate Synthase from Pseudomonas Aeruginosa
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3ZMU
 
 | | LSD1-CoREST in complex with PKSFLV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PKSFLV PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
3ZV9
 
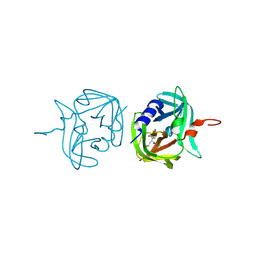 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 74 | | Descriptor: | 3C PROTEASE, ETHYL (4R)-4-[(TERT-BUTOXYCARBONYL)AMINO]-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
1RTI
 
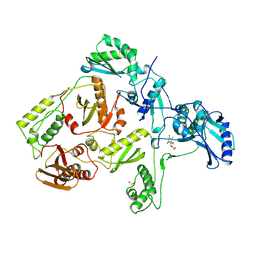 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | 1-(2-HYDROXYETHYLOXYMETHYL)-6-PHENYL THIOTHYMINE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-05-03 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
3ZO9
 
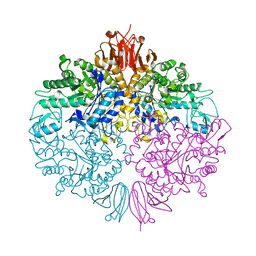 | | The structure of Trehalose Synthase (TreS) of Mycobacterium smegmatis | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Caner, S, Nguyen, N, Aguda, A, Zhang, R, Pan, Y.T, Withers, S.G, Brayer, G.D. | | Deposit date: | 2013-02-21 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The Structure of the Mycobacterium Smegmatis Trehalose Synthase Reveals an Unusual Active Site Configuration and Acarbose-Binding Mode.
Glycobiology, 23, 2013
|
|
3ZWP
 
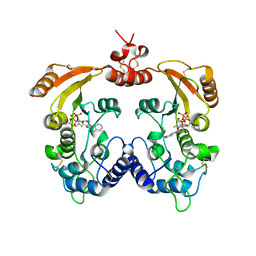 | | Crystal structure of ADP ribosyl cyclase complexed with ara-2'F-ADP- ribose at 2.1 angstrom | | Descriptor: | ADP-RIBOSYL CYCLASE, GLYCEROL, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-02 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
J.Mol.Biol., 415, 2012
|
|
6IWJ
 
 | | A designed domain swapped dimer | | Descriptor: | Archeal Protein MK0293 | | Authors: | Nandwani, N, Negi, H, Das, R. | | Deposit date: | 2018-12-05 | | Release date: | 2019-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A five-residue motif for the design of domain swapping in proteins.
Nat Commun, 10, 2019
|
|
6J1A
 
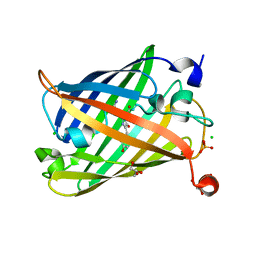 | | Photoswitchable fluorescent protein Gamillus, off-state | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Nakashima, R, Sakurai, K, shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
3ZFQ
 
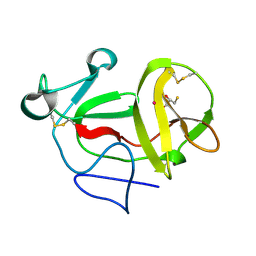 | | Crystal structure of product-like, processed N-terminal protease Npro with mercury | | Descriptor: | MERCURY (II) ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
6JEO
 
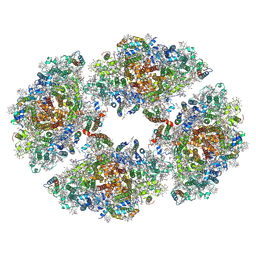 | | Structure of PSI tetramer from Anabaena | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nagao, R, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-02-06 | | Release date: | 2019-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a cyanobacterial photosystem I tetramer revealed by cryo-electron microscopy.
Nat Commun, 10, 2019
|
|
4DY4
 
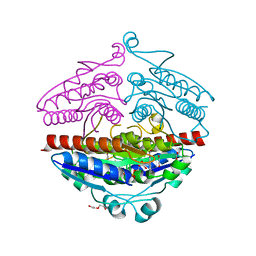 | | High resolution structure of E.coli WrbA with FMN | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, Flavoprotein wrbA | | Authors: | Kishko, I, Brynda, J, Kuta Smatanova, I, Ettrich, R, Carey, J. | | Deposit date: | 2012-02-28 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High resolution structure of E.coli WrbA with FMN
To be Published
|
|
6JJM
 
 | | Crystal structure of Mumps virus hemagglutinin-neuraminidase bound to the oligosaccharide portion of the GM2 ganglioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HN protein, ... | | Authors: | Kubota, M, Matsuoka, R, Suzuki, T, Yonekura, K, Yanagi, Y, Hashiguchi, T. | | Deposit date: | 2019-02-26 | | Release date: | 2019-05-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Molecular Mechanism of the Flexible Glycan Receptor Recognition by Mumps Virus.
J.Virol., 93, 2019
|
|
