2KKP
 
 | | Solution NMR structure of the phage integrase SAM-like Domain from Moth 1796 from Moorella thermoacetica. Northeast Structural Genomics Consortium Target MtR39K (residues 64-171). | | Descriptor: | Phage integrase | | Authors: | Ramelot, T.A, Wang, H, Ciccosanti, C, Jang, M, Nair, R, Rost, B, Swapna, G, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-26 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the phage integrase SAM-like Domain from Moth 1796
from Moorella thermoacetica. Northeast
Structural Genomics Consortium Target MtR39K (residues 64-171).
To be Published
|
|
4Z8K
 
 | |
5IP4
 
 | | X-RAY STRUCTURE OF THE C-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin, XA4551 NANOBODY AGAINST C-DCX | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
4ZNS
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with a 3-Fluoro-substituted OBHS derivative | | Descriptor: | 3-fluorophenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor-interacting peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-05 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
4ZO2
 
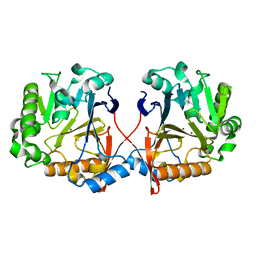 | | AidC, a Dizinc Quorum-Quenching Lactonase | | Descriptor: | Acylhomoserine lactonase, ZINC ION | | Authors: | Mascarenhas, R, Thomas, P.W, Wu, C.-X, Nocek, B.P, Hoang, Q, Fast, W, Liu, D. | | Deposit date: | 2015-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Structural and Biochemical Characterization of AidC, a Quorum-Quenching Lactonase with Atypical Selectivity.
Biochemistry, 54, 2015
|
|
4ZOL
 
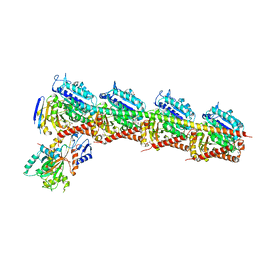 | | Crystal Structure of Tubulin-Stathmin-TTL-Tubulysin M Complex | | Descriptor: | (2R,4R)-4-{[(2-{(1R,3R)-1-(acetyloxy)-4-methyl-3-[methyl(N-{[(2S)-1-methylpiperidin-2-yl]carbonyl}-D-isoleucyl)amino]pentyl}-1,3-thiazol-4-yl)carbonyl]amino}-2-methyl-5-phenylpentanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Zhang, R. | | Deposit date: | 2015-05-06 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into the Pharmacophore of Vinca Domain Inhibitors of Microtubules
Mol.Pharmacol., 89, 2016
|
|
1HLG
 
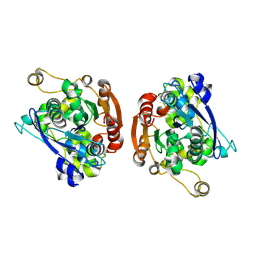 | | CRYSTAL STRUCTURE OF HUMAN GASTRIC LIPASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LIPASE, ... | | Authors: | Roussel, A, Canaan, S, Verger, R, Cambillau, C. | | Deposit date: | 1999-03-12 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human gastric lipase and model of lysosomal acid lipase, two lipolytic enzymes of medical interest.
J.Biol.Chem., 274, 1999
|
|
5I7H
 
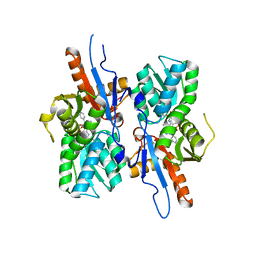 | | Mycobacterium tuberculosis CysM in complex with the Urea-scaffold inhibitor 6 [3-(3-(4-Bromophenyl)ureido)benzoic acid] | | Descriptor: | 3-{[(4-bromophenyl)carbamoyl]amino}benzoic acid, CHLORIDE ION, O-phosphoserine sulfhydrylase, ... | | Authors: | Schnell, R, Maric, S, Schneider, G. | | Deposit date: | 2016-02-17 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Inhibitors of the Cysteine Synthase CysM with Antibacterial Potency against Dormant Mycobacterium tuberculosis.
J.Med.Chem., 59, 2016
|
|
5I36
 
 | | Crystal structure of color device state A | | Descriptor: | DNA (26-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*GP*TP*GP*GP*TP*AP*TP*C)-3'), DNA (5'-D(*CP*AP*GP*AP*TP*AP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*TP*AP*CP*G)-3'), ... | | Authors: | Hao, Y, Kristiansen, M, Sha, R, Birktoft, J, Mao, C, Seeman, N.C. | | Deposit date: | 2016-02-09 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (5.123 Å) | | Cite: | A device that operates within a self-assembled 3D DNA crystal.
Nat Chem, 9, 2017
|
|
1HJM
 
 | | HUMAN PRION PROTEIN AT PH 7.0 | | Descriptor: | MAJOR PRION PROTEIN PRECURSOR | | Authors: | Calzolai, L, Zahn, R. | | Deposit date: | 2003-02-27 | | Release date: | 2003-07-03 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Influence of Ph on NMR Structure and Stability of the Human Prion Protein Globular Domain
J.Biol.Chem., 278, 2003
|
|
1HP0
 
 | | CRYSTAL STRUCTURE OF AN INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE FROM TRYPANOSOMA VIVAX IN COMPLEX WITH THE SUBSTRATE ANALOGUE 3-DEAZA-ADENOSINE | | Descriptor: | 3-DEAZA-ADENOSINE, CALCIUM ION, INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE | | Authors: | Versees, W, Decanniere, K, Pelle, R, Depoorter, J, Parkin, D.W, Steyaert, J. | | Deposit date: | 2000-12-12 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of a novel purine specific nucleoside hydrolase from Trypanosoma vivax.
J.Mol.Biol., 307, 2001
|
|
4YYU
 
 | | Ficin C crystal form I | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ficin isoform C, SULFATE ION | | Authors: | Azarkan, M, Baeyens-Volant, D, Loris, R. | | Deposit date: | 2015-03-24 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.177 Å) | | Cite: | Crystal structure of four variants of the protease Ficin from the common fig
To Be Published
|
|
4ZIA
 
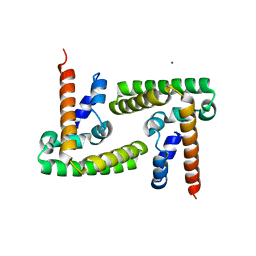 | | Crystal Structure of STAT3 N-terminal domain | | Descriptor: | FORMIC ACID, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Hu, T, Chopra, R. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Impact of the N-Terminal Domain of STAT3 in STAT3-Dependent Transcriptional Activity.
Mol.Cell.Biol., 35, 2015
|
|
4YZZ
 
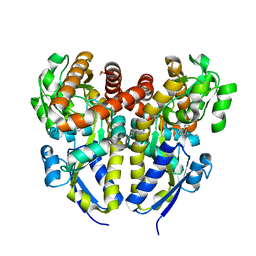 | | Crystal structure of a TRAP transporter solute binding protein (IPR025997) from Bordetella bronchiseptica RB50 (BB0280, TARGET EFI-500035) mixed occupancy dimer, copurified calcium and picolinate bound active site versus apo site | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-25 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a TRAP transporter solute binding protein (IPR025997) from Bordetella bronchiseptica RB50 (BB0280, TARGET EFI-500035) mixed occupancy dimer, copurified calcium and picolinate bound active site versus apo site
To be published
|
|
5IAA
 
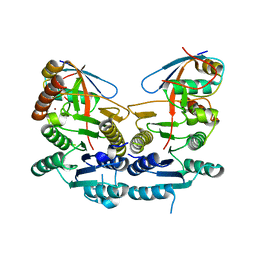 | | Crystal structure of human UBA5 in complex with UFM1 | | Descriptor: | Ubiquitin-fold modifier 1, Ubiquitin-like modifier-activating enzyme 5, ZINC ION | | Authors: | Oweis, W, Padala, P, Wiener, R. | | Deposit date: | 2016-02-21 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Trans-Binding Mechanism of Ubiquitin-like Protein Activation Revealed by a UBA5-UFM1 Complex.
Cell Rep, 16, 2016
|
|
1HNX
 
 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH PACTAMYCIN | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Brodersen, D.E, Clemons Jr, W.M, Carter, A.P, Morgan-Warren, R, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-08 | | Release date: | 2001-02-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit.
Cell(Cambridge,Mass.), 103, 2000
|
|
4Z16
 
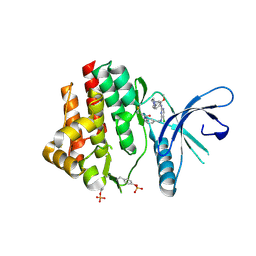 | | Crystal Structure of the Jak3 Kinase Domain Covalently Bound to N-(3-(((5-chloro-2-((2-methoxy-4-(4-methylpiperazin-1-yl)phenyl)amino)pyrimidin-4-yl)amino)methyl)phenyl)acrylamide | | Descriptor: | N-(3-{[(5-chloro-2-{[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]methyl}phenyl)prop-2-enamide, Tyrosine-protein kinase JAK3 | | Authors: | McNally, R, Tan, L, Gray, N.S, Eck, M.J. | | Deposit date: | 2015-03-26 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Development of Selective Covalent Janus Kinase 3 Inhibitors.
J.Med.Chem., 58, 2015
|
|
4ZO3
 
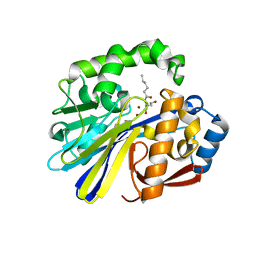 | | AidC, a Dizinc Quorum-Quenching Lactonase, in complex with a product N-hexnoyl-L-homoserine | | Descriptor: | Acylhomoserine lactonase, N-hexanoyl-L-homoserine, ZINC ION | | Authors: | Mascarenhas, R, Thomas, P.W, Wu, C.-X, Nocek, B.P, Hoang, Q, Fast, W, Liu, D. | | Deposit date: | 2015-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural and Biochemical Characterization of AidC, a Quorum-Quenching Lactonase with Atypical Selectivity.
Biochemistry, 54, 2015
|
|
5KO4
 
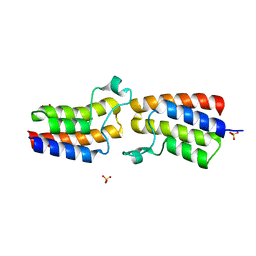 | | Bromodomain from Trypanosoma brucei Tb427.10.8150 | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | El Bakkouri, M, Walker, J.R, Hou, C.F.D, Lin, Y.H, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-29 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Bromodomain from Trypanosoma brucei Tb427.10.8150
To be published
|
|
5KPX
 
 | | Structure of RelA bound to ribosome in presence of A/R tRNA (Structure IV) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-07-05 | | Release date: | 2016-09-28 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
1HNZ
 
 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH HYGROMYCIN B | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Brodersen, D.E, Clemons Jr, W.M, Carter, A.P, Morgan-Warren, R, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-08 | | Release date: | 2001-02-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit.
Cell(Cambridge,Mass.), 103, 2000
|
|
5KQZ
 
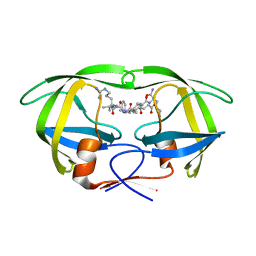 | | Protease E35D-CaP2 | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, Protease | | Authors: | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
1KG5
 
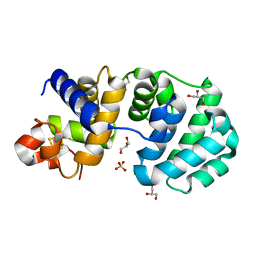 | | Crystal structure of the K142Q mutant of E.coli MutY (core fragment) | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
4Z8G
 
 | |
1HQP
 
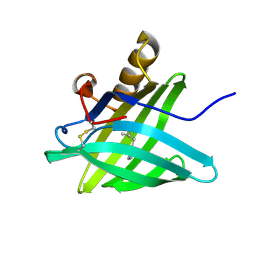 | | CRYSTAL STRUCTURE OF A TRUNCATED FORM OF PORCINE ODORANT-BINDING PROTEIN | | Descriptor: | 2-ISOBUTYL-3-METHOXYPYRAZINE, ODORANT-BINDING PROTEIN | | Authors: | Perduca, M, Mancia, F, Del Giorgio, R, Monaco, H.L. | | Deposit date: | 2000-12-19 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a truncated form of porcine odorant-binding protein.
Proteins, 42, 2001
|
|
