8BK9
 
 | |
8BKB
 
 | |
1CZ3
 
 | | DIHYDROFOLATE REDUCTASE FROM THERMOTOGA MARITIMA | | Descriptor: | DIHYDROFOLATE REDUCTASE, SULFATE ION | | Authors: | Dams, T, Auerbach, G, Bader, G, Ploom, T, Huber, R, Jaenicke, R. | | Deposit date: | 1999-09-01 | | Release date: | 2000-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of dihydrofolate reductase from Thermotoga maritima: molecular features of thermostability.
J.Mol.Biol., 297, 2000
|
|
8BL7
 
 | |
8BLD
 
 | |
8BMO
 
 | | Structure of GroEL:GroES complex exhibiting ADP-conformation in trans ring obtained under the continuous turnover conditions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, Co-chaperonin GroES, ... | | Authors: | Dhurandhar, M, Torino, S, Efremov, R. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Time-resolved cryo-EM using a combination of droplet microfluidics with on-demand jetting.
Nat.Methods, 20, 2023
|
|
8BLY
 
 | |
8BKA
 
 | |
8BKZ
 
 | | GroEL:GroES-ATP complex under continuous turnover conditions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, Co-chaperonin GroES, ... | | Authors: | Dhurandhar, M, Torino, S, Efremov, R. | | Deposit date: | 2022-11-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Time-resolved cryo-EM using a combination of droplet microfluidics with on-demand jetting.
Nat.Methods, 20, 2023
|
|
8BLE
 
 | |
1D1G
 
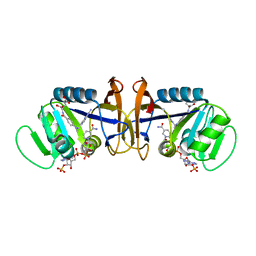 | | DIHYDROFOLATE REDUCTASE FROM THERMOTOGA MARITIMA | | Descriptor: | DIHYDROFOLATE REDUCTASE, METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Dams, T, Auerbach, G, Bader, G, Ploom, T, Huber, R, Jaenicke, R. | | Deposit date: | 1999-09-16 | | Release date: | 2000-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of dihydrofolate reductase from Thermotoga maritima: molecular features of thermostability.
J.Mol.Biol., 297, 2000
|
|
8BM1
 
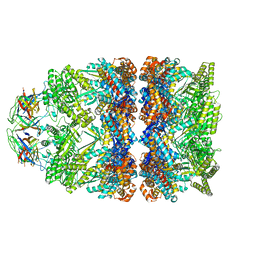 | | Structure of GroEL:GroES-ATP complex under continuous turnover conditions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, Co-chaperonin GroES, ... | | Authors: | Dhurandhar, M, Torino, S, Efremov, R. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Time-resolved cryo-EM using a combination of droplet microfluidics with on-demand jetting.
Nat.Methods, 20, 2023
|
|
8BL2
 
 | | Structure of GroEL-ATP complex plunge frozen 200 ms after reaction initiation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, MAGNESIUM ION, ... | | Authors: | Dhurandhar, M, Torino, S, Efremov, R. | | Deposit date: | 2022-11-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Time-resolved cryo-EM using a combination of droplet microfluidics with on-demand jetting.
Nat.Methods, 20, 2023
|
|
8BK7
 
 | |
8BKG
 
 | |
8BLF
 
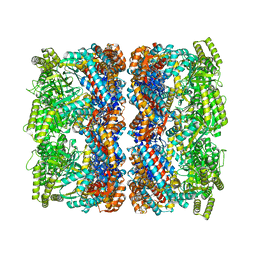 | |
8BMT
 
 | | Structure of GroEL:GroES-ATP complex plunge frozen 200 ms after reaction initiation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, Co-chaperonin GroES, ... | | Authors: | Dhurandhar, M, Efremov, R, Torino, S. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Time-resolved cryo-EM using a combination of droplet microfluidics with on-demand jetting.
Nat.Methods, 20, 2023
|
|
8BK8
 
 | |
1G9N
 
 | | HIV-1 YU2 GP120 ENVELOPE GLYCOPROTEIN COMPLEXED WITH CD4 AND INDUCED NEUTRALIZING ANTIBODY 17B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, HEAVY CHAIN, ... | | Authors: | Kwong, P.D, Wyatt, R, Majeed, S, Robinson, J, Sweet, R.W, Sodroski, J, Hendrickson, W.A. | | Deposit date: | 2000-11-25 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates.
Structure Fold.Des., 8, 2000
|
|
8BLC
 
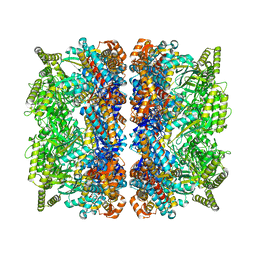 | | Structure of the GroEL-ATP complex plunge-frozen 50 ms after mixing with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, MAGNESIUM ION, ... | | Authors: | Dhurandhar, M, Torino, S, Efremov, R. | | Deposit date: | 2022-11-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Time-resolved cryo-EM using a combination of droplet microfluidics with on-demand jetting.
Nat.Methods, 20, 2023
|
|
8BMD
 
 | | Structure of GroEL-ATP complex under continuous turnover conditions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, MAGNESIUM ION, ... | | Authors: | Dhurandhar, M, Torino, S, Efremov, R. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Time-resolved cryo-EM using a combination of droplet microfluidics with on-demand jetting.
Nat.Methods, 20, 2023
|
|
8H4M
 
 | | Crystal Structure of GTP-bound Irgb6_T95D mutant | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, T-cell-specific guanine nucleotide triphosphate-binding protein 2 | | Authors: | Saijo-Hamano, Y, Okuma, H, Sakai, N, Kato, T, Imasaki, T, Nitta, R. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis of Irgb6 inactivation by Toxoplasma gondii through the phosphorylation of switch I
To Be Published
|
|
4DLM
 
 | | Crystal structure of an amidohydrolase (COG3618) from burkholderia multivorans (TARGET EFI-500235) with bound ZN, space group P212121 | | Descriptor: | Amidohydrolase 2, ZINC ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N.F, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-06 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | Crystal structure of an amidohydrolase (COG3618) from burkholderia multivorans (TARGET EFI-500235) with bound ZN, space group P212121
to be published
|
|
8HGM
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, NIV-11 Fab light chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8H4O
 
 | | Crystal Structure of nucleotide-free Irgb6_T95D mutant | | Descriptor: | T-cell-specific guanine nucleotide triphosphate-binding protein 2 | | Authors: | Saijo-Hamano, Y, Okuma, H, Sakai, N, Kato, T, Imasaki, T, Nitta, R. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-18 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of Irgb6 inactivation by Toxoplasma gondii through the phosphorylation of switch I.
Genes Cells, 29, 2024
|
|
