6KLF
 
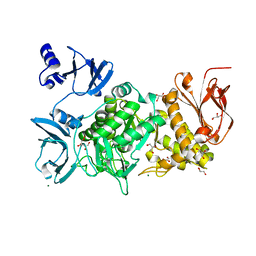 | |
4TYM
 
 | | Crystal structure of purine nucleoside phosphorylase from Streptococcus agalactiae 2603V/R, NYSGRC Target 030935 | | Descriptor: | Purine nucleoside phosphorylase DeoD-type, SULFATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-07-08 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Streptococcus agalactiae 2603V/R, NYSGRC Target 030935.
To Be Published
|
|
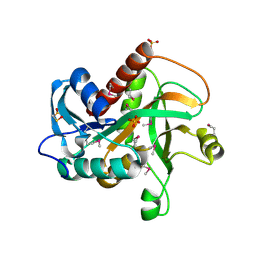
1MHN
 
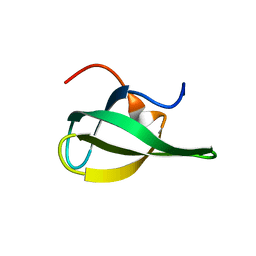 | |
1W19
 
 | | Lumazine Synthase from Mycobacterium tuberculosis bound to 3-(1,3,7- trihydro-9-D-ribityl-2,6,8-purinetrione-7-yl)propane 1-phosphate | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (4S,5S)-1,2-DITHIANE-4,5-DIOL, ... | | Authors: | Morgunova, E, Meining, W, Illarionov, B, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2004-06-03 | | Release date: | 2005-03-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Lumazine Synthase from Mycobacterium Tuberculosis as a Target for Rational Drug Design: Binding Mode of a New Class of Purinetrione Inhibitors(,)
Biochemistry, 44, 2005
|
|
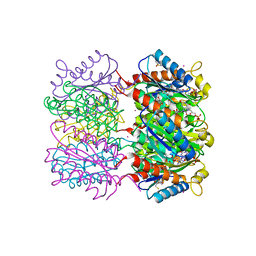
8RPZ
 
 | | Escherichia coli 50S subunit in complex with the antimicrobial peptide Api88 - conformation I | | Descriptor: | 23S ribosomal RNA, 5S ribosomal RNA, Apidaecins type 88, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C. | | Deposit date: | 2024-01-17 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Multimodal binding and inhibition of bacterial ribosomes by the antimicrobial peptides Api137 and Api88.
Nat Commun, 15, 2024
|
|
8RPY
 
 | | Escherichia coli 50S subunit in complex with the antimicrobial peptide Api137 | | Descriptor: | 23S ribosomal RNA, 5S ribosomal RNA, Apidaecins type 137, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C. | | Deposit date: | 2024-01-17 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Multimodal binding and inhibition of bacterial ribosomes by the antimicrobial peptides Api137 and Api88.
Nat Commun, 15, 2024
|
|
1W08
 
 | | STRUCTURE OF T70N HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME | | Authors: | Johnson, R, Christodoulou, J, Luisi, B, Dumoulin, M, Caddy, G, Alcocer, M, Murtagh, G, Archer, D.B, Dobson, C.M. | | Deposit date: | 2004-06-02 | | Release date: | 2004-06-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rationalising Lysozyme Amyloidosis: Insights from the Structure and Solution Dynamics of T70N Lysozyme.
J.Mol.Biol., 352, 2005
|
|
1W7F
 
 | |
8RQ0
 
 | | Escherichia coli 50S subunit in complex with the antimicrobial peptide Api88 - conformation II | | Descriptor: | 23S ribosomal RNA, 5S ribosomal RNA, Apidaecins type 88, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C. | | Deposit date: | 2024-01-17 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Multimodal binding and inhibition of bacterial ribosomes by the antimicrobial peptides Api137 and Api88.
Nat Commun, 15, 2024
|
|
1W4C
 
 | | P4 protein from Bacteriophage PHI12 apo state | | Descriptor: | NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
8RQH
 
 | | Crystal Structure of the flavoprotein monooxygenase TrlE from Streptomyces cyaneofuscatus Soc7 | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Sowa, S.T, Hoeing, L.S, Jakob, R.P, Maier, T, Teufel, R. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biosynthesis of the bacterial antibiotic 3,7-dihydroxytropolone through enzymatic salvaging of catabolic shunt products.
Chem Sci, 15, 2024
|
|
1N0V
 
 | | Crystal structure of elongation factor 2 | | Descriptor: | Elongation factor 2 | | Authors: | Joergensen, R, Ortiz, P.A, Carr-Schmid, A, Nissen, P, Kinzy, T.G, Andersen, G.R. | | Deposit date: | 2002-10-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase.
Nat.Struct.Biol., 10, 2003
|
|
6QD7
 
 | | EM structure of a EBOV-GP bound to 3T0331 neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, Envelope glycoprotein,Virion spike glycoprotein,EBOV-GP1, ... | | Authors: | Diskin, R, Cohen-Dvashi, H. | | Deposit date: | 2019-01-01 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Polyclonal and convergent antibody response to Ebola virus vaccine rVSV-ZEBOV.
Nat. Med., 25, 2019
|
|
5KKG
 
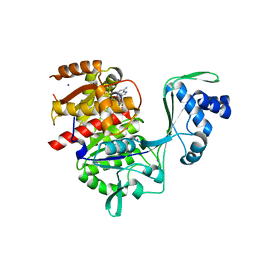 | | Crystal structure of E72A mutant of ancestral protein ancMT of ADP-dependent sugar kinases family | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, IODIDE ION, ... | | Authors: | Castro-Fernandez, V, Herrera-Morande, A, Zamora, R, Merino, F, Pereira, H.M, Brandao-Neto, J, Garratt, R, Guixe, V. | | Deposit date: | 2016-06-21 | | Release date: | 2017-07-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | Reconstructed ancestral enzymes reveal that negative selection drove the evolution of substrate specificity in ADP-dependent kinases.
J. Biol. Chem., 292, 2017
|
|
5LAS
 
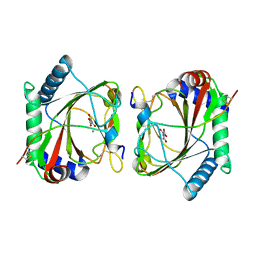 | |
6K76
 
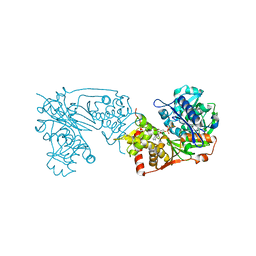 | |
7Y4N
 
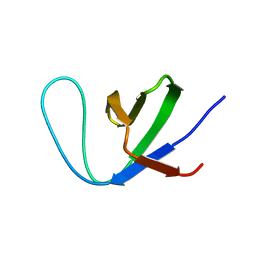 | | Insight into the C-terminal SH3 domain mediated binding of Drosophila Drk to Sos and Dos | | Descriptor: | Growth factor receptor-bound protein 2 | | Authors: | Pooppadi, M.S, Ikeya, T, Sugasawa, H, Watanabe, R, Mishima, M, Inomata, K, Ito, Y. | | Deposit date: | 2022-06-15 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insight into the C-terminal SH3 domain mediated binding of Drosophila Drk to Sos and Dos.
Biochem.Biophys.Res.Commun., 625, 2022
|
|
7QA1
 
 | | The structure of natural crystals of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein elucidated using serial femtosecond crystallography at an X-ray free electron laser | | Descriptor: | Toxin-10 pesticidal protein (Tpp) 49Aa1 | | Authors: | Williamson, L.J, Rizkallah, P.J, Berry, C, Oberthur, D, Galchenkova, M, Yefanov, O, Bean, R, Best, H.L. | | Deposit date: | 2021-11-15 | | Release date: | 2023-05-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein elucidated from natural crystals using MHz-SFX.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7XXF
 
 | | Structure of photosynthetic LH1-RC super-complex of Rhodopila globiformis | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (6~{E},8~{E},10~{E},12~{E},14~{E},16~{E},18~{E},20~{E},22~{E},24~{E},26~{E},28~{E})-2,31-dimethoxy-2,6,10,14,19,23,27,31-octamethyl-dotriaconta-6,8,10,12,14,16,18,20,22,24,26,28-dodecaen-5-one, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Tani, K, Kanno, R, Kurosawa, K, Takaichi, S, Nagashima, K.V.P, Hall, M, Yu, L.-J, Kimura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Wang-Otomo, Z.-Y. | | Deposit date: | 2022-05-30 | | Release date: | 2022-11-16 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | An LH1-RC photocomplex from an extremophilic phototroph provides insight into origins of two photosynthesis proteins.
Commun Biol, 5, 2022
|
|
6KP1
 
 | | Crystal structure of two domain M1 zinc metallopeptidase E323A mutant bound to L-methionine amino acid | | Descriptor: | METHIONINE, SODIUM ION, ZINC ION, ... | | Authors: | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | Deposit date: | 2019-08-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis for the unusual substrate specificity of unique two-domain M1 metallopeptidase.
Int.J.Biol.Macromol., 147, 2020
|
|
8DQA
 
 | |
8DQ6
 
 | |
6KP0
 
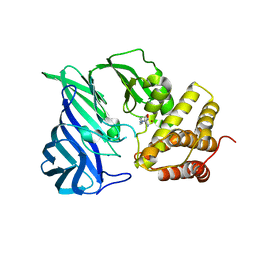 | | Crystal structure of two domain M1 zinc metallopeptidase E323A mutant bound to L-arginine | | Descriptor: | ARGININE, SODIUM ION, ZINC ION, ... | | Authors: | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the unusual substrate specificity of unique two-domain M1 metallopeptidase.
Int.J.Biol.Macromol., 147, 2020
|
|
6L40
 
 | | Discovery of novel peptidomimetic boronate ClpP inhibitors with noncanonical enzyme mechanism as potent virulence blockers in vitro and in vivo | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, [(1S)-3-methyl-1-[[(2S)-3-phenyl-2-(pyrazin-2-ylcarbonylamino)propanoyl]amino]butyl]boronic acid | | Authors: | Luo, Y.F, Bao, R, Ju, Y, He, L.H. | | Deposit date: | 2019-10-15 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Discovery of Novel Peptidomimetic Boronate ClpP Inhibitors with Noncanonical Enzyme Mechanism as Potent Virulence Blockersin Vitroandin Vivo.
J.Med.Chem., 63, 2020
|
|
6KOZ
 
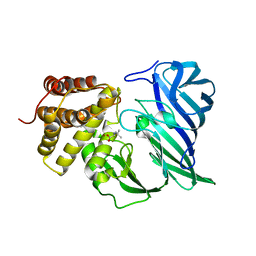 | | Crystal structure of two domain M1 zinc metallopeptidase E323 mutant bound to L-Leucine amino acid | | Descriptor: | LEUCINE, SODIUM ION, ZINC ION, ... | | Authors: | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the unusual substrate specificity of unique two-domain M1 metallopeptidase.
Int.J.Biol.Macromol., 147, 2020
|
|
