6S76
 
 | | Crystal structure of human Nek7 | | 分子名称: | DI(HYDROXYETHYL)ETHER, Serine/threonine-protein kinase Nek7 | | 著者 | Nasir, N, Bayliss, R. | | 登録日 | 2019-07-04 | | 公開日 | 2020-06-10 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.38 Å) | | 主引用文献 | Nek7 conformational flexibility and inhibitor binding probed through protein engineering of the R-spine.
Biochem.J., 477, 2020
|
|
5EIJ
 
 | | Carbonic Anhydrase II in complex with Sulfonamide Inhibitor | | 分子名称: | 1-(3-iodanylphenyl)-3-(4-sulfamoylphenyl)thiourea, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | 著者 | Lomelino, C.L, Mahon, B.P, McKenna, R. | | 登録日 | 2015-10-29 | | 公開日 | 2016-11-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Kinetic and X-ray crystallographic investigations on carbonic anhydrase isoforms I, II, IX and XII of a thioureido analog of SLC-0111.
Bioorg. Med. Chem., 24, 2016
|
|
5ERW
 
 | |
6RLO
 
 | | Crystal structure of AT1412dm Fab fragment in complex with CD9 large extracellular loop | | 分子名称: | AT1412dm Fab Fragment (Heavy Chain), AT1412dm Fab Fragment (Light Chain), CD9 antigen, ... | | 著者 | Neviani, V, Pearce, N.M, Pos, W, Schotte, R, Spits, H, Gros, P. | | 登録日 | 2019-05-02 | | 公開日 | 2021-05-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis of a homo-dimerization site in tetraspanin CD9 targeted by a melanoma patient-derived antibody
To Be Published
|
|
6RLM
 
 | | Crystal structure of AT1412dm Fab fragment | | 分子名称: | AT1412dm Fab (Heavy Chain), AT1412dm Fab (Light Chain), CHLORIDE ION | | 著者 | Neviani, V, Pearce, N.M, Pos, W, Schotte, R, Spits, H, Gros, P. | | 登録日 | 2019-05-02 | | 公開日 | 2021-05-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis of a homo-dimerization site in tetraspanin CD9 targeted by a melanoma patient-derived antibody
To Be Published
|
|
6S0N
 
 | | A9 peptide derived from Herceptin fab binding region | | 分子名称: | GLN-ASP-VAL-ASN-THR-ALA-VAL-ALA-TRP | | 著者 | De Luca, S, Verdoliva, V, Saviano, M, Fattorusso, R, Diana, D. | | 登録日 | 2019-06-17 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | SPR and NMR characterization of the molecular interaction between A9 peptide and a model system of HER2 receptor: A fragment approach for selecting peptide structures specific for their target.
J.Pept.Sci., 26, 2020
|
|
6S8J
 
 | | Structure of ZEBOV GP in complex with 5T0180 antibody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope Glycoprotein 1, Envelope glycoprotein, ... | | 著者 | Diskin, R, Cohen-Dvashi, H. | | 登録日 | 2019-07-10 | | 公開日 | 2020-02-12 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (2.91 Å) | | 主引用文献 | Structural Basis for a Convergent Immune Response against Ebola Virus.
Cell Host Microbe, 27, 2020
|
|
6S9A
 
 | | Artificial GTPase-BSE dimer of human Dynamin1 | | 分子名称: | CHLORIDE ION, Dynamin-1,Dynamin-1, ZINC ION | | 著者 | Ganichkin, O.M, Vancraenenbroeck, R, Rosenblum, G, Hofmann, H, Daumke, O, Noel, J.K. | | 登録日 | 2019-07-11 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Quantification and demonstration of the collective constriction-by-ratchet mechanism in the dynamin molecular motor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2YPW
 
 | | Atomic model for the N-terminus of TraO fitted in the full-length structure of the bacterial pKM101 type IV secretion system core complex | | 分子名称: | TRAO | | 著者 | Rivera-Calzada, A, Fronzes, R, Savva, C.G, Chandran, V, Lian, P.W, Laeremans, T, Pardon, E, Steyaert, J, Remaut, H, Waksman, G, Orlova, E.V. | | 登録日 | 2012-11-02 | | 公開日 | 2013-04-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (12.4 Å) | | 主引用文献 | Structure of a Bacterial Type Iv Secretion Core Complex at Subnanometre Resolution.
Embo J., 32, 2013
|
|
5EO8
 
 | | Crystal structure of AOL(868) | | 分子名称: | Predicted protein, methyl 1-seleno-beta-L-fucopyranoside | | 著者 | Kato, R, Kiso, M, Ishida, H, Ando, H, Suzuki, T, Shimabukuro, S, Makyio, H. | | 登録日 | 2015-11-10 | | 公開日 | 2016-06-29 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Six independent fucose-binding sites in the crystal structure of Aspergillus oryzae lectin
Biochem.Biophys.Res.Commun., 477, 2016
|
|
5ENE
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with 5-Amino-2-benzyl-1,3-oxazole-4-carbonitrile (SGC - Diamond I04-1 fragment screening) | | 分子名称: | 5-azanyl-2-(phenylmethyl)-1,3-oxazole-4-carbonitrile, PH-interacting protein | | 著者 | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Amin, J, Szykowska, A, Burgess-Brown, N, Spencer, J, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | 登録日 | 2015-11-09 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
6SH3
 
 | |
3O20
 
 | | Electron transfer complexes:experimental mapping of the Redox-dependent Cytochrome C electrostatic surface | | 分子名称: | Cytochrome c, HEME C, NITRATE ION | | 著者 | De March, M, De Zorzi, R, Casini, A, Messori, L, Geremia, S, Demitri, N, Gabbiani, C, Guerri, A. | | 登録日 | 2010-07-22 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Nitrate as a probe of cytochrome c surface: crystallographic identification of crucial "hot spots" for protein-protein recognition.
J. Inorg. Biochem., 135, 2014
|
|
5F4C
 
 | | Crystal Structure of Ribonuclease Inhibitor Barstar from Salmonella Typhimurium | | 分子名称: | MALONATE ION, Putative cytoplasmic protein | | 著者 | Maltseva, N, Kim, Y, Mulligan, R, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-12-03 | | 公開日 | 2015-12-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal Structure of Ribonuclease Inhibitor Barstar from Salmonella Typhimurium.
To Be Published
|
|
6S8I
 
 | | Structure of ZEBOV GP in complex with 3T0265 antibody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, Enveloped Glycoprotein 1, ... | | 著者 | Diskin, R, Cohen-Dvashi, H. | | 登録日 | 2019-07-10 | | 公開日 | 2020-02-12 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Structural Basis for a Convergent Immune Response against Ebola Virus.
Cell Host Microbe, 27, 2020
|
|
5F6W
 
 | | Crystal structure of Ubc9 (K48/K49A/E54A) complexed with Fragment 1 (biphenol) | | 分子名称: | 2-(2-hydroxyphenyl)phenol, SUMO-conjugating enzyme UBC9 | | 著者 | Lountos, G.T, Hewitt, W.M, Zlotkowski, K, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | 登録日 | 2015-12-07 | | 公開日 | 2016-04-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.699 Å) | | 主引用文献 | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5F73
 
 | | Crystal structure of Mutant S12T of Adenosine/Methylthioadenosine Phosphorylase in APO form | | 分子名称: | Methylthioadenosine phosphorylase, SULFATE ION | | 著者 | Torini, J.R.S, Brandao-Neto, J, DeMarco, R, Pereira, H.M. | | 登録日 | 2015-12-07 | | 公開日 | 2016-12-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Crystal Structure of Schistosoma mansoni Adenosine Phosphorylase/5'-Methylthioadenosine Phosphorylase and Its Importance on Adenosine Salvage Pathway.
PLoS Negl Trop Dis, 10, 2016
|
|
5F1C
 
 | | Crystal structure of an invertebrate P2X receptor from the Gulf Coast tick in the presence of ATP and Zn2+ ion at 2.9 Angstroms | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, Putative uncharacterized protein, ... | | 著者 | Kasuya, G, Hattori, M, Ishitani, R, Nureki, O. | | 登録日 | 2015-11-30 | | 公開日 | 2016-03-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural Insights into Divalent Cation Modulations of ATP-Gated P2X Receptor Channels
Cell Rep, 14, 2016
|
|
6SF6
 
 | |
6SCW
 
 | |
5EZR
 
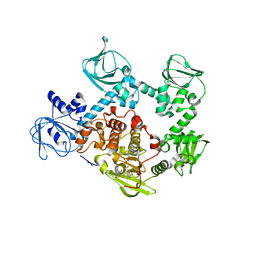 | | Crystal Structure of PVX_084705 bound to compound | | 分子名称: | CHLORIDE ION, N-[5-(3-{2-[(cyclopropylmethyl)amino]pyrimidin-4-yl}-7-[(dimethylamino)methyl]-6-methylimidazo[1,2-a]pyridin-2-yl)-2-fluorophenyl]methanesulfonamide, cGMP-dependent protein kinase, ... | | 著者 | El Bakkouri, M, Amani, M, Walker, J.R, Osborne, S, Large, J.M, Birchall, K, Bouloc, N, Smiljanic-Hurley, E, Wheldon, M, Harding, D.J, Merritt, A.T, Ansell, K.H, Coombs, P.J, Kettleborough, C.A, Stewart, B.L, Bowyer, P.W, Gutteridge, W.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Baker, D.A, Hui, R, Loppnau, P, Structural Genomics Consortium (SGC) | | 登録日 | 2015-11-26 | | 公開日 | 2017-05-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of PVX_084705 bound to compound
To Be Published
|
|
3NZP
 
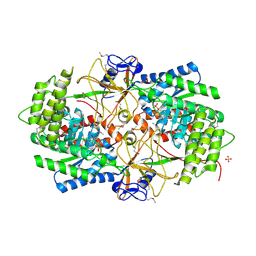 | | Crystal Structure of the Biosynthetic Arginine decarboxylase SpeA from Campylobacter jejuni, Northeast Structural Genomics Consortium Target BR53 | | 分子名称: | Arginine decarboxylase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Forouhar, F, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-07-16 | | 公開日 | 2010-09-01 | | 最終更新日 | 2012-02-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structures of bacterial biosynthetic arginine decarboxylases.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
6SGK
 
 | |
5F6V
 
 | | Crystal structure of Ubc9 (K48/K49A/E54A) complexed with Fragment 1 (biphenol from fragment cocktail screen) | | 分子名称: | 2-(2-hydroxyphenyl)phenol, SUMO-conjugating enzyme UBC9 | | 著者 | Lountos, G.T, Hewitt, W.M, Zlotkowski, K, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | 登録日 | 2015-12-07 | | 公開日 | 2016-04-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.492 Å) | | 主引用文献 | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5F77
 
 | | Crystal structure of Mutant S12T of adenosine/Methylthioadenosine phosphorylase from Schistosoma mansoni in complex with Adenine | | 分子名称: | ADENINE, Methylthioadenosine phosphorylase, SULFATE ION | | 著者 | Torini, J.R.S, Brandao-Neto, J, DeMarco, R, Pereira, H.M. | | 登録日 | 2015-12-07 | | 公開日 | 2016-12-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Crystal Structure of Schistosoma mansoni Adenosine Phosphorylase/5'-Methylthioadenosine Phosphorylase and Its Importance on Adenosine Salvage Pathway.
PLoS Negl Trop Dis, 10, 2016
|
|
