8EG4
 
 | | [iU:Ag+:iU] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | 分子名称: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*(5IU)P*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*(5IU)P*AP*CP*A*(AG))-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | 著者 | Vecchioni, S, Lu, B, Seeman, N.C, Sha, R, Ohayon, Y.P. | | 登録日 | 2022-09-10 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (4.18 Å) | | 主引用文献 | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 35, 2023
|
|
5GNV
 
 | | Structure of PSD-95/MAP1A complex reveals unique target recognition mode of MAGUK GK domain | | 分子名称: | Disks large homolog 4, Microtubule-associated protein 1A, SULFATE ION | | 著者 | Shang, Y, Xia, Y, Zhu, R, Zhu, J. | | 登録日 | 2016-07-25 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.596 Å) | | 主引用文献 | Structure of the PSD-95/MAP1A complex reveals a unique target recognition mode of the MAGUK GK domain
Biochem. J., 474, 2017
|
|
6YQ5
 
 | | Hybrid structure of the SPP1 tail tube by solid-state NMR and cryo EM - NMR Ensemble | | 分子名称: | Tail tube protein gp17.1* | | 著者 | Zinke, M, Sachowsky, K.A.A, Zinn-Justin, S, Ravelli, R, Schroder, G.F, Habeck, M, Lange, A. | | 登録日 | 2020-04-16 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (4 Å), SOLID-STATE NMR | | 主引用文献 | Architecture of the flexible tail tube of bacteriophage SPP1.
Nat Commun, 11, 2020
|
|
5GQW
 
 | |
6YVW
 
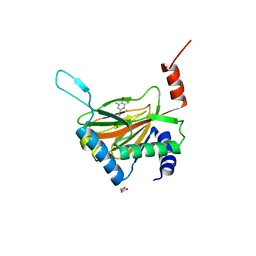 | | HIF prolyl hydroxylase 2 (PHD2/ EGLN1) in complex with monocyclic BB-328 | | 分子名称: | 4-[(5-bromanyl-4,6-dimethyl-pyridin-2-yl)amino]-4-oxidanylidene-butanoic acid, Egl nine homolog 1, FE (III) ION, ... | | 著者 | Chowdhury, R, Banerji, B, Schofield, C.J. | | 登録日 | 2020-04-28 | | 公開日 | 2020-12-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Use of cyclic peptides to induce crystallization: case study with prolyl hydroxylase domain 2.
Sci Rep, 10, 2020
|
|
8D8O
 
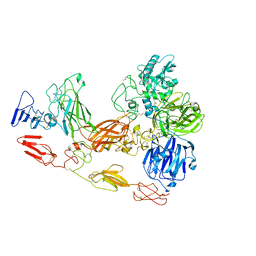 | | Cryo-EM structure of substrate unbound PAPP-A | | 分子名称: | Pappalysin-1, ZINC ION | | 著者 | Judge, R.A, Jain, R, Hao, Q, Ouch, C, Sridar, J, Smith, C.L, Wang, J.C.K, Eaton, D. | | 登録日 | 2022-06-08 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Structure of the PAPP-ABP5 complex reveals mechanism of substrate recognition
Nat Commun, 13, 2022
|
|
6YW1
 
 | |
5GQD
 
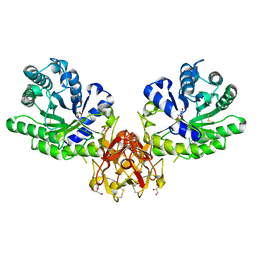 | | Crystal structure of covalent glycosyl-enzyme intermediate of xylanase mutant (T82A, N127S, and E128H) from Streptomyces olivaceoviridis E-86 | | 分子名称: | Beta-xylanase, GLYCEROL, beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | 著者 | Suzuki, R, Fujimoto, Z, Kaneko, S, Kuno, A. | | 登録日 | 2016-08-07 | | 公開日 | 2017-08-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Azidolysis by the Formation of Stable Ser-His Catalytic Dyad in a Glycoside Hydrolase Family 10 Xylanase Mutant
J.Appl.Glyosci., 65, 2019
|
|
5GQZ
 
 | |
6THU
 
 | |
2ZIO
 
 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in complex with AlocLys-AMP and PNP | | 分子名称: | 5'-O-[(S)-({(2S)-2-amino-6-[(propoxycarbonyl)amino]hexanoyl}oxy)(hydroxy)phosphoryl]adenosine, IMIDODIPHOSPHORIC ACID, Pyrrolysyl-tRNA synthetase | | 著者 | Yanagisawa, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2008-02-19 | | 公開日 | 2008-12-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Multistep Engineering of Pyrrolysyl-tRNA Synthetase to Genetically Encode N(varepsilon)-(o-Azidobenzyloxycarbonyl) lysine for Site-Specific Protein Modification
Chem.Biol., 15, 2008
|
|
8DDU
 
 | | cryo-EM structure of TRPM3 ion channel in the presence of PIP2, state3 | | 分子名称: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | 著者 | Zhao, C, MacKinnon, R. | | 登録日 | 2022-06-19 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDQ
 
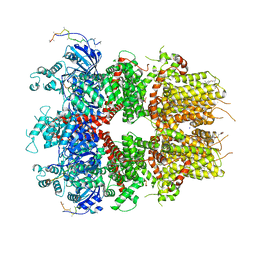 | | cryo-EM structure of TRPM3 ion channel in the presence of soluble Gbg, focused on channel | | 分子名称: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | 著者 | Zhao, C, MacKinnon, R. | | 登録日 | 2022-06-18 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
1IMD
 
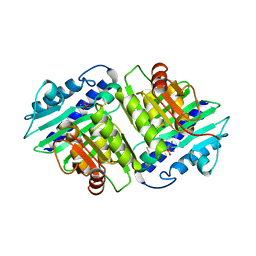 | |
8DDS
 
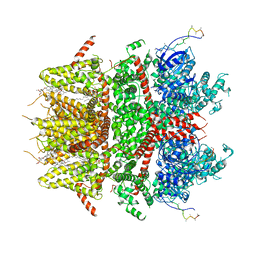 | | cryo-EM structure of TRPM3 ion channel in the presence of PIP2, state1 | | 分子名称: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | 著者 | Zhao, C, MacKinnon, R. | | 登録日 | 2022-06-18 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDW
 
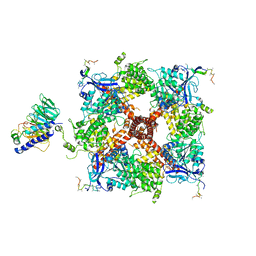 | |
3UGV
 
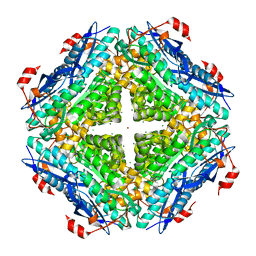 | | Crystal structure of an enolase from alpha pretobacterium bal199 (EFI TARGET EFI-501650) with bound MG | | 分子名称: | CHLORIDE ION, Enolase, MAGNESIUM ION, ... | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2011-11-03 | | 公開日 | 2011-11-23 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of an enolase from alpha pretobacterium bal199 (EFI TARGET EFI-501650) with bound MG
to be published
|
|
8DDT
 
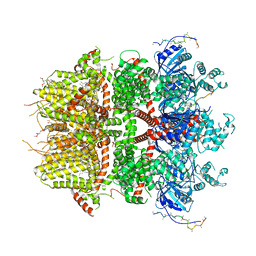 | | cryo-EM structure of TRPM3 ion channel in the presence of PIP2, state2 | | 分子名称: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | 著者 | Zhao, C, MacKinnon, R. | | 登録日 | 2022-06-18 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDR
 
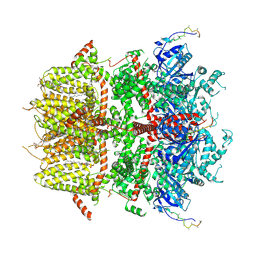 | | cryo-EM structure of TRPM3 ion channel in the absence of PIP2 | | 分子名称: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | 著者 | Zhao, C, MacKinnon, R. | | 登録日 | 2022-06-18 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDX
 
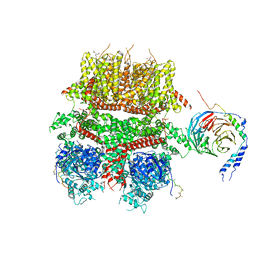 | | cryo-EM structure of TRPM3 ion channel in complex with Gbg in the presence of PIP2, tethered by ALFA-nanobody | | 分子名称: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Zhao, C, MacKinnon, R. | | 登録日 | 2022-06-19 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8DDV
 
 | | Cryo-EM structure of TRPM3 ion channel in the presence of PIP2, state4 | | 分子名称: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | 著者 | Zhao, C, MacKinnon, R. | | 登録日 | 2022-06-19 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
6YVZ
 
 | | HIF prolyl hydroxylase 2 (PHD2/ EGLN1) in complex with bicyclic JLS-367 | | 分子名称: | 4-[(5-bromanylisoquinolin-3-yl)amino]-4-oxidanylidene-butanoic acid, BICARBONATE ION, Egl nine homolog 1, ... | | 著者 | Chowdhury, R, Sorensen, J.L, Schofield, C.J. | | 登録日 | 2020-04-29 | | 公開日 | 2020-12-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Use of cyclic peptides to induce crystallization: case study with prolyl hydroxylase domain 2.
Sci Rep, 10, 2020
|
|
8DJ9
 
 | | Carbonic Anhydrase II in complex with Ibuprofen | | 分子名称: | (2R)-2-[4-(2-methylpropyl)phenyl]propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Carbonic anhydrase 2, ... | | 著者 | Combs, J.C, McKenna, R, Andring, J.T. | | 登録日 | 2022-06-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Ibuprofen: a weak inhibitor of carbonic anhydrase II.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
6YW0
 
 | |
1IUQ
 
 | | The 1.55 A Crystal Structure of Glycerol-3-Phosphate Acyltransferase | | 分子名称: | GLYCEROL, Glycerol-3-Phosphate Acyltransferase, SULFATE ION | | 著者 | Tamada, T, Feese, M.D, Kato, Y, Kuroki, R. | | 登録日 | 2002-03-06 | | 公開日 | 2003-10-07 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Substrate recognition and selectivity of plant glycerol-3-phosphate acyltransferases (GPATs) from Cucurbita moscata and Spinacea oleracea.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
